Hypoxia inducible lncRNA-CBSLR modulates ferroptosis through m6A-YTHDF2-dependent modulation of CBS in gastric cancer
- PMID: 35499052
- PMCID: PMC9039740
- DOI: 10.1016/j.jare.2021.10.001
Hypoxia inducible lncRNA-CBSLR modulates ferroptosis through m6A-YTHDF2-dependent modulation of CBS in gastric cancer
Abstract
Introduction: Tumors are usually refractory to anti-cancer therapeutics under hypoxic conditions. However, the underlying molecular mechanism remains to be elucidated.
Objectives: Our study intended to identify hypoxia inducible lncRNAs and their biological function in gastric cancer (GC).
Methods: Differentially expressed lncRNAs were determined by microarray analysis between GC cells exposed to hypoxia (1% O2) and normoxia (21% O2) for 24 h. The expression level of CBSLR was manipulated in several GC cell lines to perform molecular and biological analyses both in vitro and in vivo.
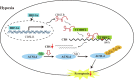
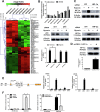
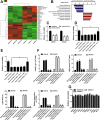
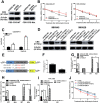
Results: We identified a hypoxia-induced lncRNA-CBSLR that protected GC cells from ferroptosis, leading to chem-resistance. Mechanically, CBSLR interacted with YTHDF2 to form a CBSLR/YTHDF2/CBS signaling axis that decreased the stability of CBS mRNA by enhancing the binding of YTHDF2 with the m6A-modified coding sequence (CDS) of CBS mRNA. Furthermore, under decreased CBS levels, the methylation of the ACSL4 protein was reduced, leading to protein polyubiquitination and degradation of ACSL4. This, in turn, decreased the pro-ferroptosis phosphatidylethanolamine (PE) (18:0/20:4) and PE (18:0/22:4) content and contributed to ferroptosis resistance. Notably, CBSLR is upregulated, whereas CBS is downregulated in GC tissues compared to matched normal tissues; and GC patients with high CBSLR/low CBS levels have a worse clinical outcome and a poorer response to chemotherapy.
Conclusion: Our study reveals a novel mechanism in how HIF1α/CBSLR modulates ferroptosis/chemoresistance in GC, illuminating potential therapeutic targets for refractory hypoxic tumors.
Keywords: 4-HNE, 4-hydroxynonenal; AJCC, American Joint Committee on Cancer; CDS, coding sequence; CHIP, chromatin immunoprecipitation; Chemoresistance; DZNeP, 3-deazaneplanocin A; FBS, fetal bovine serum; Ferroptosis; GC, gastric cancer; GEO, Gene Expression Omnibus; Gastric cancer; HE, Hematoxylin and eosin; HREs, hypoxia response elements; Hypoxia; IHC, immunohistochemical; MDA, malondialdehyde; RIP, RNA immunoprecipitation; TCGA, the cancer genome atlas; TNM, tumor-node-metastasis staging system; YTHDF2, YTH domain family protein 2; lncRNA; lncRNA, long noncoding RNA; qPCR, quantitative real-time PCR.
© 2022 The Authors. Published by Elsevier B.V. on behalf of Cairo University.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. - PubMed
-
- Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. - PubMed
-
- Smyth E.C., Nilsson M., Grabsch H.I., van Grieken N.CT., Lordick F. Smyth, Magnus Nilsson, Heike I Grabsch, Nicole Ct van Grieken Florian Lordick. Gastric cancer. Lancet. 2020;396(10251):635–648. - PubMed
-
- Bhandari V., Hoey C., Liu L.Y., Lalonde E., Ray J., Livingstone J., et al. Molecular landmarks of tumor hypoxia across cancer types. Nat Genet. 2019;51(2):308–318. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

