Activated SUMOylation restricts MHC class I antigen presentation to confer immune evasion in cancer
- PMID: 35499080
- PMCID: PMC9057585
- DOI: 10.1172/JCI152383
Activated SUMOylation restricts MHC class I antigen presentation to confer immune evasion in cancer
Abstract
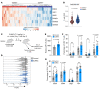
Activated SUMOylation is a hallmark of cancer. Starting from a targeted screening for SUMO-regulated immune evasion mechanisms, we identified an evolutionarily conserved function of activated SUMOylation, which attenuated the immunogenicity of tumor cells. Activated SUMOylation allowed cancer cells to evade CD8+ T cell-mediated immunosurveillance by suppressing the MHC class I (MHC-I) antigen-processing and presentation machinery (APM). Loss of the MHC-I APM is a frequent cause of resistance to cancer immunotherapies, and the pharmacological inhibition of SUMOylation (SUMOi) resulted in reduced activity of the transcriptional repressor scaffold attachment factor B (SAFB) and induction of the MHC-I APM. Consequently, SUMOi enhanced the presentation of antigens and the susceptibility of tumor cells to CD8+ T cell-mediated killing. Importantly, SUMOi also triggered the activation of CD8+ T cells and thereby drove a feed-forward loop amplifying the specific antitumor immune response. In summary, we showed that activated SUMOylation allowed tumor cells to evade antitumor immunosurveillance, and we have expanded the understanding of SUMOi as a rational therapeutic strategy for enhancing the efficacy of cancer immunotherapies.
Keywords: Antigen presentation; Immunology; Immunotherapy; Oncology; Ubiquitin-proteosome system.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

