Tn FLXopen: Markerless Transposons for Functional Fluorescent Fusion Proteins and Protein Interaction Prediction
- PMID: 35499319
- PMCID: PMC9241775
- DOI: 10.1128/spectrum.02428-21
Tn FLXopen: Markerless Transposons for Functional Fluorescent Fusion Proteins and Protein Interaction Prediction
Abstract
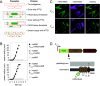
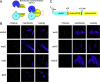
Fluorescence microscopy of cells expressing proteins translationally linked to a fluorophore can be a powerful tool to investigate protein localization dynamics in vivo. One major obstacle to reliably analyze biologically relevant localization is the construction of a fusion protein that is both fluorescent and functional. Here, we develop a strategy to construct fluorescent fusions at theoretically any location in the protein by using TnFLXopen random transposon mutagenesis to randomly insert a gene encoding a fluorescent protein. Moreover, insertions within a target gene are enriched by an inducible gene-trap strategy and selection by fluorescence activated cell sorting. Using this approach, we isolate a variety of fluorescent fusions to FtsZ that exhibit ring-like localization and a fusion to the flagellar stator protein that both is functional for supporting motility and localizes as fluorescent puncta. Finally, we further modify TnFLXopen to insert the coding sequence for the C-terminal half of mVenus for use in bimolecular fluorescence complementation (BiFC) and the in vivo detection of protein-protein interaction candidates. As proof-of-concept, the DivIVA polar scaffolding protein was fused to the N terminus of mVenus, the C terminus of mVenus was delivered by transposition, and a combination of fluorescence activated cell sorter (FACS) sorting and whole-genome sequencing identified the known self-interaction of DivIVA as well as other possible candidate interactors. We suggest that the FACS selection is a viable alternative to antibiotic selection in transposon mutagenesis that can generate new fluorescent tools for in vivo protein characterization. IMPORTANCE Transposon mutagenesis is a powerful tool for random mutagenesis, as insertion of a transposon and accompanying antibiotic resistance cassette often disrupt gene function. Here, we present a series of transposons with fluorescent protein genes which, when integrated in frame, may be selected with a fluorescence activated cell sorter (FACS). An open reading frame runs continuously through the transposon such that fluorescent protein fusions may be inserted theoretically anywhere in the primary sequence and potentially preserve function of the target protein. Finally, the transposons were further modified to randomly insert a partial fluorescent protein compatible with bimolecular fluorescence complementation (BiFC) to identify protein interaction candidates.
Keywords: Bacillus subtilis; BiFC; DivIVA; FACS; FtsZ; MotA; internal fluorescent protein fusion; protein-protein interactions; transposon mutagenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
New transposons to generate GFP protein fusions in Candida albicans.Gene. 2008 Jul 1;417(1-2):13-8. doi: 10.1016/j.gene.2008.03.005. Epub 2008 Mar 19. Gene. 2008. PMID: 18467040
-
Construction and validation of the Tn5-PLtetO-1-msfGFP transposon as a tool to probe protein expression and localization.J Microbiol Methods. 2019 Jun;161:56-62. doi: 10.1016/j.mimet.2019.04.012. Epub 2019 Apr 18. J Microbiol Methods. 2019. PMID: 31004623
-
Libraries of green fluorescent protein fusions generated by transposition in vitro.Gene. 1998 Nov 19;222(2):213-22. doi: 10.1016/s0378-1119(98)00503-4. Gene. 1998. PMID: 9831655
-
Applications of transposon-based gene delivery system in bacteria.J Microbiol Biotechnol. 2009 Mar;19(3):217-28. doi: 10.4014/jmb.0811.669. J Microbiol Biotechnol. 2009. PMID: 19349746 Review.
-
Transposon-based strategies for microbial functional genomics and proteomics.Annu Rev Genet. 2003;37:3-29. doi: 10.1146/annurev.genet.37.110801.142807. Annu Rev Genet. 2003. PMID: 14616054 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

