A CLEIA Antigen Assay in Diagnosis and Follow-Up of SARS-CoV-2-Positive Subjects
- PMID: 35499325
- PMCID: PMC9241685
- DOI: 10.1128/spectrum.01032-21
A CLEIA Antigen Assay in Diagnosis and Follow-Up of SARS-CoV-2-Positive Subjects
Abstract
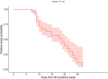
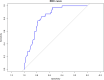
This study includes 259 consecutive nasopharyngeal swabs which tested positive for a molecular SARS-CoV-2 test and 77 subjects who were followed longitudinally, with nasopharyngeal swabs performed weekly until clinical recovery and a negative result for the molecular test were reached. All swabs were also tested with a Lumipulse SARS-CoV-2 chemiluminescence enzyme immunoassay (CLEIA) antigen assay. The antigen test was positive in 169 (65.3%) out of the 259 subjects, while no antigen was detected in 90 subjects (34.7%). In the antigen-positive subjects, clinical status moved slightly toward a more frequent presence of symptoms. Longitudinal follow-up shows how the time of negativization has a faster kinetic in the antigenic test than in the molecular test. Antigenic test result values, considered as a time-dependent covariate and log-transformed, were highly associated with the time to negative swab, with good prediction ability. Receiver operating characteristic (ROC) curve analysis showed a very good discrimination ability of antigenic tests in classifying negative swabs. The optimal cutoff which jointly maximized sensitivity and specificity was 1.55, resulting in an overall accuracy of 0.75, a sensitivity of 0.73, and a specificity of 0.83. After dichotomizing the antigenic test according to the previously determined cutoff value of 1.55, the time-dependent covariate Cox model again suggests a highly significant association of antigenic test values with the time to negative swab molecular: a subject with an antigenic test value lower than 1.55 had almost a 13-fold higher probability to also result negative in the molecular test compared to a subject with an antigenic test value higher than 1.55. IMPORTANCE Our work explores the possibility of using a sensible and reliable antigenic test in a wider range of SARS-CoV-2 diagnostic and clinical applications. Furthermore, this tool seems particularly promising in follow-up with infected subjects, because while the molecular test frequently yields the persistence of low positivities, raising yet unanswered questions, this antigenic test shows more uniform and faster negativization during the evolution of the infection, somehow paralleling the dynamics of infectivity. Although more data will be required to definitely prove it, we believe these findings might be of great interest.
Keywords: SARS-CoV-2; antigenic test; nasopharyngeal swabs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Johns Hopkins University (JHU), Center for Systems Science and Engineering (CSSE). COVID-19 dashboard. JHU, Baltimore, MD. Available at: https://coronavirus.jhu.edu/map.html. Accessed July 7th, 2021.
-
- Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, Xie G, Lin S, Wang R, Yang X, Chen W, Wang Q, Zhang D, Liu Y, Gong R, Ma Z, Lu S, Xiao Y, Gu Y, Zhang J, Yao H, Xu K, Lu X, Wei G, Zhou J, Fang Q, Cai H, Qiu Y, Sheng J, Chen Y, Liang T. 2020. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China. January–March 2020: retrospective cohort study. BMJ 369:m1443. doi: 10.1136/bmj.m1443. - DOI - PMC - PubMed
-
- Cohen AN, Kessel B, Milgroom MG. 2020. Diagnosing COVID-19 infection: the danger of over-reliance on positive test results. medRxiv. doi: 10.1101/2020.04.26.20080911. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

