Making genomic surveillance deliver: A lineage classification and nomenclature system to inform rabies elimination
- PMID: 35500026
- PMCID: PMC9162366
- DOI: 10.1371/journal.ppat.1010023
Making genomic surveillance deliver: A lineage classification and nomenclature system to inform rabies elimination
Abstract
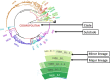
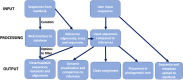
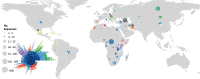
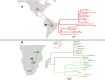
The availability of pathogen sequence data and use of genomic surveillance is rapidly increasing. Genomic tools and classification systems need updating to reflect this. Here, rabies virus is used as an example to showcase the potential value of updated genomic tools to enhance surveillance to better understand epidemiological dynamics and improve disease control. Previous studies have described the evolutionary history of rabies virus, however the resulting taxonomy lacks the definition necessary to identify incursions, lineage turnover and transmission routes at high resolution. Here we propose a lineage classification system based on the dynamic nomenclature used for SARS-CoV-2, defining a lineage by phylogenetic methods for tracking virus spread and comparing sequences across geographic areas. We demonstrate this system through application to the globally distributed Cosmopolitan clade of rabies virus, defining 96 total lineages within the clade, beyond the 22 previously reported. We further show how integration of this tool with a new rabies virus sequence data resource (RABV-GLUE) enables rapid application, for example, highlighting lineage dynamics relevant to control and elimination programmes, such as identifying importations and their sources, as well as areas of persistence and routes of virus movement, including transboundary incursions. This system and the tools developed should be useful for coordinating and targeting control programmes and monitoring progress as countries work towards eliminating dog-mediated rabies, as well as having potential for broader application to the surveillance of other viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- WHO | Rabies. [cited 12 Apr 2021]. Available: https://www.who.int/news-room/fact-sheets/detail/rabies
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

