Tolypocladamide H and the Proposed Tolypocladamide NRPS in Tolypocladium Species
- PMID: 35500108
- PMCID: PMC9150700
- DOI: 10.1021/acs.jnatprod.2c00153
Tolypocladamide H and the Proposed Tolypocladamide NRPS in Tolypocladium Species
Abstract
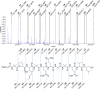
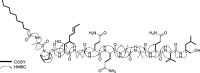
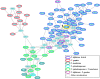
The genome of entomopathogenic fungus Tolypocladium inflatum Gams encodes 43 putative biosynthetic gene clusters for specialized metabolites, although genotype-phenotype linkages have been reported only for the cyclosporins and fumonisins. T. inflatum was cultured in defined minimal media, supplemented with or without one of nine different amino acids. Acquisition of LC-MS/MS data for molecular networking and manual analysis facilitated annotation of putative known and unknown metabolites. These data led us to target a family of peptaibols and guided the isolation and purification of tolypocladamide H (1), which showed modest antibacterial activity and toxicity to mammalian cells at micromolar concentrations. HRMS/MS, NMR, and advanced Marfey's analysis were used to assign the structure of 1 as a peptaibol containing 4-[(E)-2-butenyl]-4-methyl-l-threonine (Bmt), a hallmark structural motif of the cyclosporins. LC-MS detection of homologous tolypocladamide metabolites and phylogenomic analyses of peptaibol biosynthetic genes in other cultured Tolypocladium species allowed assignment of a putative tolypocladamide nonribosomal peptide synthetase gene.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

