Overlapping Delta and Omicron Outbreaks During the COVID-19 Pandemic: Dynamic Panel Data Estimates
- PMID: 35500140
- PMCID: PMC9169703
- DOI: 10.2196/37377
Overlapping Delta and Omicron Outbreaks During the COVID-19 Pandemic: Dynamic Panel Data Estimates
Abstract
Background: The Omicron variant of SARS-CoV-2 is more transmissible than prior variants of concern (VOCs). It has caused the largest outbreaks in the pandemic, with increases in mortality and hospitalizations. Early data on the spread of Omicron were captured in countries with relatively low case counts, so it was unclear how the arrival of Omicron would impact the trajectory of the pandemic in countries already experiencing high levels of community transmission of Delta.
Objective: The objective of this study is to quantify and explain the impact of Omicron on pandemic trajectories and how they differ between countries that were or were not in a Delta outbreak at the time Omicron occurred.
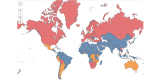
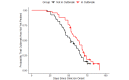
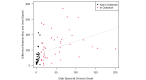
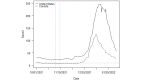
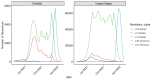
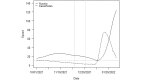
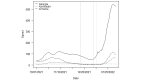
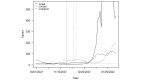
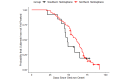
Methods: We used SARS-CoV-2 surveillance and genetic sequence data to classify countries into 2 groups: those that were in a Delta outbreak (defined by at least 10 novel daily transmissions per 100,000 population) when Omicron was first sequenced in the country and those that were not. We used trend analysis, survival curves, and dynamic panel regression models to compare outbreaks in the 2 groups over the period from November 1, 2021, to February 11, 2022. We summarized the outbreaks in terms of their peak rate of SARS-CoV-2 infections and the duration of time the outbreaks took to reach the peak rate.
Results: Countries that were already in an outbreak with predominantly Delta lineages when Omicron arrived took longer to reach their peak rate and saw greater than a twofold increase (2.04) in the average apex of the Omicron outbreak compared to countries that were not yet in an outbreak.
Conclusions: These results suggest that high community transmission of Delta at the time of the first detection of Omicron was not protective, but rather preluded larger outbreaks in those countries. Outbreak status may reflect a generally susceptible population, due to overlapping factors, including climate, policy, and individual behavior. In the absence of strong mitigation measures, arrival of a new, more transmissible variant in these countries is therefore more likely to lead to larger outbreaks. Alternately, countries with enhanced surveillance programs and incentives may be more likely to both exist in an outbreak status and detect more cases during an outbreak, resulting in a spurious relationship. Either way, these data argue against herd immunity mitigating future outbreaks with variants that have undergone significant antigenic shifts.
Keywords: Arellano-Bond estimator; B.1.1.529; COVID-19; Delta; Omicron variant of concern; SARS-CoV-2; disease transmission metrics; dynamic panel data; outbreak; stringency index; surveillance.
©Alexander L Lundberg, Ramon Lorenzo-Redondo, Judd F Hultquist, Claudia A Hawkins, Egon A Ozer, Sarah B Welch, P V Vara Prasad, Chad J Achenbach, Janine I White, James F Oehmke, Robert L Murphy, Robert J Havey, Lori A Post. Originally published in JMIR Public Health and Surveillance (https://publichealth.jmir.org), 03.06.2022.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures
Similar articles
-
Has Omicron Changed the Evolution of the Pandemic?JMIR Public Health Surveill. 2022 Jan 31;8(1):e35763. doi: 10.2196/35763. JMIR Public Health Surveill. 2022. PMID: 35072638 Free PMC article.
-
SARS-CoV-2 Wave Two Surveillance in East Asia and the Pacific: Longitudinal Trend Analysis.J Med Internet Res. 2021 Feb 1;23(2):e25454. doi: 10.2196/25454. J Med Internet Res. 2021. PMID: 33464207 Free PMC article.
-
Updated Surveillance Metrics and History of the COVID-19 Pandemic (2020-2023) in East Asia and the Pacific Region: Longitudinal Trend Analysis.JMIR Public Health Surveill. 2025 Feb 21;11:e53214. doi: 10.2196/53214. JMIR Public Health Surveill. 2025. PMID: 39804185 Free PMC article.
-
Is the SARS CoV-2 Omicron Variant Deadlier and More Transmissible Than Delta Variant?Int J Environ Res Public Health. 2022 Apr 11;19(8):4586. doi: 10.3390/ijerph19084586. Int J Environ Res Public Health. 2022. PMID: 35457468 Free PMC article. Review.
-
The outbreak of SARS-CoV-2 Omicron lineages, immune escape, and vaccine effectivity.J Med Virol. 2023 Jan;95(1):e28138. doi: 10.1002/jmv.28138. Epub 2022 Sep 21. J Med Virol. 2023. PMID: 36097349 Free PMC article. Review.
Cited by
-
COVID-19 Pandemic Risk Assessment: Systematic Review.Risk Manag Healthc Policy. 2024 Apr 11;17:903-925. doi: 10.2147/RMHP.S444494. eCollection 2024. Risk Manag Healthc Policy. 2024. PMID: 38623576 Free PMC article. Review.
-
South Asia's COVID-19 History and Surveillance: Updated Epidemiological Assessment.JMIR Public Health Surveill. 2024 Aug 26;10:e53331. doi: 10.2196/53331. JMIR Public Health Surveill. 2024. PMID: 39013116 Free PMC article.
-
SARS-CoV-2 Omicron Variant Genomic Sequences and Their Epidemiological Correlates Regarding the End of the Pandemic: In Silico Analysis.JMIR Bioinform Biotechnol. 2023 Jan 10;4:e42700. doi: 10.2196/42700. eCollection 2023. JMIR Bioinform Biotechnol. 2023. PMID: 36688013 Free PMC article.
References
-
- Kandeel M, Mohamed MEM, Abd El-Lateef HM, Venugopala KN, El-Beltagi HS. Omicron variant genome evolution and phylogenetics. J Med Virol. 2022 Apr 15;94(4):1627–1632. doi: 10.1002/jmv.27515. http://europepmc.org/abstract/MED/34888894 - DOI - PMC - PubMed
-
- Viana R, Moyo S, Amoako DG, Tegally H, Scheepers C, Althaus CL, Anyaneji UJ, Bester PA, Boni MF, Chand M, Choga WT, Colquhoun R, Davids M, Deforche K, Doolabh D, du Plessis L, Engelbrecht S, Everatt J, Giandhari J, Giovanetti M, Hardie D, Hill V, Hsiao N-Y, Iranzadeh A, Ismail A, Joseph C, Joseph R, Koopile L, Kosakovsky Pond SL, Kraemer MUG, Kuate-Lere L, Laguda-Akingba O, Lesetedi-Mafoko O, Lessells RJ, Lockman S, Lucaci AG, Maharaj A, Mahlangu B, Maponga T, Mahlakwane K, Makatini Z, Marais G, Maruapula D, Masupu K, Matshaba M, Mayaphi S, Mbhele N, Mbulawa MB, Mendes A, Mlisana K, Mnguni A, Mohale T, Moir M, Moruisi K, Mosepele M, Motsatsi G, Motswaledi MS, Mphoyakgosi T, Msomi N, Mwangi PN, Naidoo Y, Ntuli N, Nyaga M, Olubayo L, Pillay S, Radibe B, Ramphal Y, Ramphal U, San JE, Scott L, Shapiro R, Singh L, Smith-Lawrence P, Stevens W, Strydom A, Subramoney K, Tebeila N, Tshiabuila D, Tsui J, van Wyk S, Weaver S, Wibmer CK, Wilkinson E, Wolter N, Zarebski AE, Zuze B, Goedhals D, Preiser W, Treurnicht F, Venter M, Williamson C, Pybus OG, Bhiman J, Glass A, Martin DP, Rambaut A, Gaseitsiwe S, von Gottberg A, de Oliveira T. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022 Mar;603(7902):679–686. doi: 10.1038/s41586-022-04411-y. http://europepmc.org/abstract/MED/35042229 10.1038/s41586-022-04411-y - DOI - PMC - PubMed
-
- VanBlargan L, Errico J, Halfmann P, Zost S, Crowe J, Purcell L, Kawaoka Y, Corti D, Fremont D, Diamond M. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med. 2021 Dec 27;:1–6. doi: 10.21203/rs.3.rs-1175516/v1. doi: 10.21203/rs.3.rs-1175516/v1. - DOI - DOI - PMC - PubMed
-
- Poudel S, Ishak A, Perez-Fernandez J, Garcia E, León-Figueroa DA, Romaní L, Bonilla-Aldana DK, Rodriguez-Morales AJ. Highly mutated SARS-CoV-2 Omicron variant sparks significant concern among global experts - what is known so far? Travel Med Infect Dis. 2022 Jan;45:102234. doi: 10.1016/j.tmaid.2021.102234. http://europepmc.org/abstract/MED/34896326 S1477-8939(21)00275-1 - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

