Variation in blood microbial lipopolysaccharide (LPS) contributes to immune reconstitution in response to suppressive antiretroviral therapy in HIV
- PMID: 35500539
- PMCID: PMC9065923
- DOI: 10.1016/j.ebiom.2022.104037
Variation in blood microbial lipopolysaccharide (LPS) contributes to immune reconstitution in response to suppressive antiretroviral therapy in HIV
Abstract
Background: In HIV infection, even under long-term antiretroviral therapy (ART), up to 20% of HIV-infected individuals fail to restore CD4+ T cell counts to the levels similar to those of healthy controls. The mechanisms of poor CD4+ T cell reconstitution on suppressive ART are not fully understood.
Methods: Here, we tested the hypothesis that lipopolysaccharide (LPS) from bacteria enriched in the plasma from immune non-responders (INRs) contributes to blunted CD4+ T cell recovery on suppressive ART in HIV. We characterized plasma microbiome in HIV INRs (aviremic, CD4+ T cell counts < 350 cells/μl), immune responders (IRs, CD4+ T cell counts > 500 cells/μl), and healthy controls. Next, we analyzed the structure of the lipid A domain of three bacterial species identified by mass spectrometry (MS) and evaluated the LPS function through LPS induced proinflammatory responses and CD4+ T cell apoptosis in PBMCs. In comparison, we also evaluated plasma levels of proinflammatory cytokine and chemokine patterns in these three groups. At last, to study the causality of microbiome-blunted CD4+ T cell recovery in HIV, B6 mice were intraperitoneally (i.p.) injected with heat-killed Burkholderia fungorum, Serratia marcescens, or Phyllobacterium myrsinacearum, twice per week for total of eight weeks.
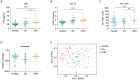
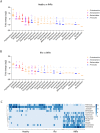
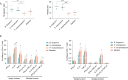
Findings: INRs exhibited elevated plasma levels of total microbial translocation compared to the IRs and healthy controls. The most enriched bacteria were Burkholderia and Serratia in INRs and were Phyllobacterium in IRs. Further, unlike P. myrsinacearum LPS, B. fungorum and S. marcescens LPS induced proinflammatory responses and CD4+ T cell apoptosis in PBMCs, and gene profiles of bacteria-mediated cell activation pathways in THP-1 cells in vitro. Notably, LPS structural analysis by mass spectrometry revealed that lipid A from P. myrsinacearum exhibited a divergent structure consistent with weak toll-like receptor (TLR) 4 agonism, similar to the biological profile of probiotic bacteria. In contrast, lipid A from B. fungorum and S. marcescens showed structures more consistent with canonical TLR4 agonists stemming from proinflammatory bacterial strains. Finally, intraperitoneal (i.p.) injection of inactivated B. fungorum and S. marcescens but not P. myrsinacearum resulted in cell apoptosis in mesenteric lymph nodes of C57BL/6 mice in vivo.
Interpretation: These results suggest that the microbial products are causally associated with INR phenotype. In summary, variation in blood microbial LPS immunogenicity may contribute to immune reconstitution in response to suppressive ART. Collectively, this work is consistent with immunologically silencing microbiome being causal and targetable with therapy in HIV.
Funding: This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID; R01 AI128864, Jiang) (NIAID; P30 AI027767, Saag/Health), the Medical Research Service at the Ralph H. Johnson VA Medical Center (merit grant VA CSRD MERIT I01 CX-002422, Jiang), and the National Institute of Aging (R21 AG074331, Scott). The SCOPE cohort was supported by the UCSF/Gladstone Institute of Virology & Immunology CFAR (P30 AI027763, Gandhi) and the CFAR Network of Integrated Clinical Systems (R24 AI067039, Saag). The National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001450 (the pilot grant, Jiang). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Keywords: HIV; Immune non-responders; Immune responders; Lipid A; Lipopolysaccharide.
Published by Elsevier B.V.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Similar articles
-
Impaired immune reconstitution in HIV infection: the role of CD4+ T-cell-associated NKG2D ligands, CD4+ T-cell subsets imbalance, and immune function deficiency.Front Immunol. 2025 Feb 21;16:1541574. doi: 10.3389/fimmu.2025.1541574. eCollection 2025. Front Immunol. 2025. PMID: 40061947 Free PMC article.
-
The predictive role of CD4+ cell count and CD4/CD8 ratio in immune reconstitution outcome among HIV/AIDS patients receiving antiretroviral therapy: an eight-year observation in China.BMC Immunol. 2019 Aug 28;20(1):31. doi: 10.1186/s12865-019-0311-2. BMC Immunol. 2019. PMID: 31455209 Free PMC article.
-
Higher proportions of circulating CXCR3+ CCR6- Tfh cells as a hallmark of impaired CD4+ T-cell recovery in HIV-1-infected immunological non-responders.mBio. 2025 May 14;16(5):e0057525. doi: 10.1128/mbio.00575-25. Epub 2025 Mar 25. mBio. 2025. PMID: 40130906 Free PMC article.
-
Alterations in circulating markers in HIV/AIDS patients with poor immune reconstitution: Novel insights from microbial translocation and innate immunity.Front Immunol. 2022 Oct 17;13:1026070. doi: 10.3389/fimmu.2022.1026070. eCollection 2022. Front Immunol. 2022. PMID: 36325329 Free PMC article. Review.
-
Incomplete immune reconstitution in HIV/AIDS patients on antiretroviral therapy: Challenges of immunological non-responders.J Leukoc Biol. 2020 Apr;107(4):597-612. doi: 10.1002/JLB.4MR1019-189R. Epub 2020 Jan 22. J Leukoc Biol. 2020. PMID: 31965635 Free PMC article. Review.
Cited by
-
Viewpoint: Is lipopolysaccharide a hormone or a vitamin?Brain Behav Immun. 2023 Nov;114:1-2. doi: 10.1016/j.bbi.2023.07.018. Epub 2023 Jul 28. Brain Behav Immun. 2023. PMID: 37517741 Free PMC article. Review. No abstract available.
-
Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update for 2021-2022.Mass Spectrom Rev. 2025 May-Jun;44(3):213-453. doi: 10.1002/mas.21873. Epub 2024 Jun 24. Mass Spectrom Rev. 2025. PMID: 38925550 Free PMC article. Review.
-
Analysis of the clinical characteristics of poor immunological reconstitution in AIDS patients after long-term antiviral therapy in Xinjiang, China.BMC Immunol. 2025 Jul 4;26(1):48. doi: 10.1186/s12865-025-00732-5. BMC Immunol. 2025. PMID: 40615782 Free PMC article.
-
The CD4/CD8 ratio is associated with T lymphocyte functions in long-term virally suppressed patients with HIV.BMC Infect Dis. 2025 Jan 17;25(1):76. doi: 10.1186/s12879-025-10469-6. BMC Infect Dis. 2025. PMID: 39825235 Free PMC article.
-
Increased Microbial Translocation is a Prognostic Biomarker of Different Immune Responses to ART in People Living with HIV.Infect Drug Resist. 2023 Jun 17;16:3871-3878. doi: 10.2147/IDR.S404384. eCollection 2023. Infect Drug Resist. 2023. PMID: 37351382 Free PMC article.
References
-
- Gazzola L., Tincati C., Bellistri G.M., Monforte A., Marchetti G. The absence of CD4+ T cell count recovery despite receipt of virologically suppressive highly active antiretroviral therapy: clinical risk, immunological gaps, and therapeutic options. Clin Infect Dis. 2009;48(3):328–337. - PubMed
-
- Lewden C., Chene G., Morlat P., et al. HIV-infected adults with a CD4 cell count greater than 500 cells/mm3 on long-term combination antiretroviral therapy reach same mortality rates as the general population. J Acquir Immune Defic Syndr. 2007;46(1):72–77. - PubMed
-
- Gutierrez F., Padilla S., Masia M., et al. Patients' characteristics and clinical implications of suboptimal CD4 T-cell gains after 1 year of successful antiretroviral therapy. Curr HIV Res. 2008;6(2):100–107. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

