In silico evidence of antiviral activity against SARS-CoV-2 main protease of oligosaccharides from Porphyridium sp
- PMID: 35500710
- PMCID: PMC9052773
- DOI: 10.1016/j.scitotenv.2022.155580
In silico evidence of antiviral activity against SARS-CoV-2 main protease of oligosaccharides from Porphyridium sp
Abstract
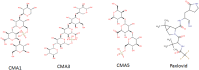
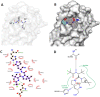
The coronavirus pandemic (COVID-19) has created an urgent need to develop effective strategies for prevention and treatment. In this context, therapies against protease Mpro, a conserved viral target, would be essential to contain the spread of the virus and reduce mortality. Using combined techniques of structure modelling, in silico docking and pharmacokinetics prediction, many compounds from algae were tested for their ability to inhibit the SARS-CoV-2 main protease and compared to the recent recognized drug Paxlovid. The screening of 27 algal molecules including 15 oligosaccharides derived from sulfated and non-sulphated polysaccharides, eight pigments and four poly unsaturated fatty acids showed high affinities to interact with the protein active site. Best candidates showing high docking scores in comparison with the reference molecule were sulfated tri-, tetra- and penta-saccharides from Porphyridium sp. exopolysaccharides (SEP). Structural and energetic analyses over 100 ns MD simulation demonstrated high SEP fragments-Mpro complex stability. Pharmacokinetics predictions revealed the prospects of the identified molecules as potential drug candidates.
Keywords: Docking; Inhibitors; Microalgae; Polysaccharide; Porphyridium; SARS-CoV-2.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

