A Large-Scale Genome-Wide Gene-Gene Interaction Study of Lung Cancer Susceptibility in Europeans With a Trans-Ethnic Validation in Asians
- PMID: 35500836
- PMCID: PMC9512697
- DOI: 10.1016/j.jtho.2022.04.011
A Large-Scale Genome-Wide Gene-Gene Interaction Study of Lung Cancer Susceptibility in Europeans With a Trans-Ethnic Validation in Asians
Abstract
Introduction: Although genome-wide association studies have been conducted to investigate genetic variation of lung tumorigenesis, little is known about gene-gene (G × G) interactions that may influence the risk of non-small cell lung cancer (NSCLC).
Methods: Leveraging a total of 445,221 European-descent participants from the International Lung Cancer Consortium OncoArray project, Transdisciplinary Research in Cancer of the Lung and UK Biobank, we performed a large-scale genome-wide G × G interaction study on European NSCLC risk by a series of analyses. First, we used BiForce to evaluate and rank more than 58 billion G × G interactions from 340,958 single-nucleotide polymorphisms (SNPs). Then, the top interactions were further tested by demographically adjusted logistic regression models. Finally, we used the selected interactions to build lung cancer screening models of NSCLC, separately, for never and ever smokers.
Results: With the Bonferroni correction, we identified eight statistically significant pairs of SNPs, which predominantly appeared in the 6p21.32 and 5p15.33 regions (e.g., rs521828C6orf10 and rs204999PRRT1, ORinteraction = 1.17, p = 6.57 × 10-13; rs3135369BTNL2 and rs2858859HLA-DQA1, ORinteraction = 1.17, p = 2.43 × 10-13; rs2858859HLA-DQA1 and rs9275572HLA-DQA2, ORinteraction = 1.15, p = 2.84 × 10-13; rs2853668TERT and rs62329694CLPTM1L, ORinteraction = 0.73, p = 2.70 × 10-13). Notably, even with much genetic heterogeneity across ethnicities, three pairs of SNPs in the 6p21.32 region identified from the European-ancestry population remained significant among an Asian population from the Nanjing Medical University Global Screening Array project (rs521828C6orf10 and rs204999PRRT1, ORinteraction = 1.13, p = 0.008; rs3135369BTNL2 and rs2858859HLA-DQA1, ORinteraction = 1.11, p = 5.23 × 10-4; rs3135369BTNL2 and rs9271300HLA-DQA1, ORinteraction = 0.89, p = 0.006). The interaction-empowered polygenetic risk score that integrated classical polygenetic risk score and G × G information score was remarkable in lung cancer risk stratification.
Conclusions: Important G × G interactions were identified and enriched in the 5p15.33 and 6p21.32 regions, which may enhance lung cancer screening models.
Keywords: Cancer risk; GWAS; Gene-gene interaction; Genetic screening model; Lung cancer; Single nucleotide polymorphism.
Copyright © 2022 International Association for the Study of Lung Cancer. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure: The authors declare no conflict of interest.
Figures


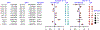

Comment in
-
Genie Out of the Bottle: Is There a Role for Gene-Gene Interactions in Early Detection of Lung Cancer?J Thorac Oncol. 2022 Aug;17(8):946-948. doi: 10.1016/j.jtho.2022.05.012. J Thorac Oncol. 2022. PMID: 35931422 No abstract available.
-
Gene-Gene Interaction in Ever Smokers With Lung Cancer: Is There Confounding by Chronic Obstructive Pulmonary Disease in Genome-Wide Association Studies?J Thorac Oncol. 2023 Mar;18(3):e23-e24. doi: 10.1016/j.jtho.2022.08.013. J Thorac Oncol. 2023. PMID: 36842813 No abstract available.
-
Reply to Scott et al: "Gene-Gene interaction in ever-smokers with lung cancer: Is there confounding by COPD in GWAS?".J Thorac Oncol. 2023 Mar;18(3):e24-e26. doi: 10.1016/j.jtho.2022.12.002. J Thorac Oncol. 2023. PMID: 36842814 No abstract available.
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. - PubMed
Publication types
MeSH terms
Grants and funding
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- P30 ES000002/ES/NIEHS NIH HHS/United States
- P30 CA076292/CA/NCI NIH HHS/United States
- R01 AG056764/AG/NIA NIH HHS/United States
- R01 CA249096/CA/NCI NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- U01 CA209414/CA/NCI NIH HHS/United States
- 001/WHO_/World Health Organization/International
- R01 CA092824/CA/NCI NIH HHS/United States
- R35 CA197449/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- U01 CA167462/CA/NCI NIH HHS/United States
- U19 CA203654/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

