Semi-nested RT-PCR enables sensitive and high-throughput detection of SARS-CoV-2 based on melting analysis
- PMID: 35500877
- PMCID: PMC9052777
- DOI: 10.1016/j.cca.2022.04.997
Semi-nested RT-PCR enables sensitive and high-throughput detection of SARS-CoV-2 based on melting analysis
Abstract
Background: Asymptomatic transmission was found to be the Achilles' heel of the symptom-based screening strategy, necessitating the implementation of mass testing to efficiently contain the transmission of COVID-19 pandemic. However, the global shortage of molecular reagents and the low throughput of available realtime PCR facilities were major limiting factors.
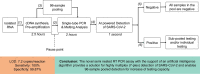
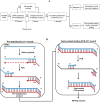
Methods: A novel semi-nested and heptaplex (7-plex) RT-PCR assay with melting analysis for detection of SARS-CoV-2 RNA has been established for either individual testing or 96-sample pooled testing. The complex melting spectrum collected from the heptaplex RT-PCR amplicons was interpreted with the support of an artificial intelligence algorithm for the detection of SARS-CoV-2 RNA. The analytical and clinical performance of the semi-nested RT-PCR assay was evaluated using RNAs synthesized in-vitro and those isolated from nasopharyngeal samples.
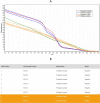
Results: The LOD of the assay for individual testing was estimated to be 7.2 copies/reaction. Clinical performance evaluation indicated a sensitivity of 100% (95% CI: 97.83-100) and a specificity of 99.87% (95% CI: 99.55-99.98). More importantly, the assay supports a breakthrough sample pooling method, which makes possible parallel screening of up to 96 samples in one real-time PCR well without loss of sensitivity. As a result, up to 8,820 individual pre-amplified samples could be screened for SARS-CoV-2 within each 96-well plate of realtime PCR using the pooled testing procedure.
Conclusion: The novel semi-nested RT-PCR assay provides a solution for highly multiplex (7-plex) detection of SARS-CoV-2 and enables 96-sample pooled detection for increase of testing capacity. .
Keywords: Artificial intelligent; COVID-19; High-throughput PCR; Melting analysis; Pooling; Semi-nested.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Authors state no conflict of interest.
The corresponding author of current manuscript, Tho Huu Ho, is also the inventor of the pending patent application (PCT/IB2021/052120), which described the novel method of pooling after the pre-amplification step for high-throughput molecular testing.
Vietnam Military Medical University - VMMU (Hanoi, Vietnam) and Mien Dong Sai Gon Clinics - MDSC (Dong Nai, Vietnam) provide major funding for this work, and are the co-applicants of mentioned pending patent application.
VMMU and MDSC had granted Ampharco U.S.A (Dong Nai, Vietnam) to use the pending application for manufacturing of the test kits (named “AmphaBio HT-HiThroughput PCR COVID-19 kit”), which then were approved by Vietnam Ministry of Health on May 7th, 2021 for emergency use in response to COVID-19 pandemic situation.
Figures
Similar articles
-
Clinical evaluation of a multiplex real-time RT-PCR assay for detection of SARS-CoV-2 in individual and pooled upper respiratory tract samples.Arch Virol. 2021 Sep;166(9):2551-2561. doi: 10.1007/s00705-021-05148-1. Epub 2021 Jul 14. Arch Virol. 2021. PMID: 34259914 Free PMC article.
-
A semi-automated, isolation-free, high-throughput SARS-CoV-2 reverse transcriptase (RT) loop-mediated isothermal amplification (LAMP) test.Sci Rep. 2021 Nov 1;11(1):21385. doi: 10.1038/s41598-021-00827-0. Sci Rep. 2021. PMID: 34725400 Free PMC article.
-
Saliva as a testing specimen with or without pooling for SARS-CoV-2 detection by multiplex RT-PCR test.PLoS One. 2021 Feb 23;16(2):e0243183. doi: 10.1371/journal.pone.0243183. eCollection 2021. PLoS One. 2021. PMID: 33621263 Free PMC article.
-
Evaluation of conventional and point-of-care real-time RT-PCR tests for the detection of SARS-CoV-2 through a pooled testing strategy.J Clin Lab Anal. 2022 Jun;36(6):e24491. doi: 10.1002/jcla.24491. Epub 2022 May 9. J Clin Lab Anal. 2022. PMID: 35535393 Free PMC article.
-
Strategies for Scaling up SARS-CoV-2 Molecular Testing Capacity.Clin Lab Med. 2022 Jun;42(2):261-282. doi: 10.1016/j.cll.2022.02.006. Epub 2022 Mar 8. Clin Lab Med. 2022. PMID: 35636826 Free PMC article. Review.
Cited by
-
Detection of novel coronaviruses from dusky fruit bat (Penthetor lucasi) in Sarawak, Malaysian Borneo.Vet Med Sci. 2023 Nov;9(6):2634-2641. doi: 10.1002/vms3.1251. Epub 2023 Sep 2. Vet Med Sci. 2023. PMID: 37658663 Free PMC article.
References
-
- Wang B., Goh Y.S., Prince T., Ngoh E.Z.X., Salleh S.N.M., Hor P.X., Loh C.Y., Fong S.W., Hartley C., Tan S.-Y., Young B.E., Leo Y.-S., Lye D.C., Maurer-Stroh S., Ng L.F.P., Hiscox J.A., Renia L., Wang C.-I. Resistance of SARS-CoV-2 variants to neutralization by convalescent plasma from early COVID-19 outbreak in Singapore. NPJ Vacc. 2021;6(1) doi: 10.1038/s41541-021-00389-2. - DOI - PMC - PubMed
-
- Zhao S., Lin Q., Ran J., Musa S.S., Yang G., Wang W., Lou Y., Gao D., Yang L., He D., Wang M.H. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int. J. Infect. Dis. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. - DOI - PMC - PubMed
-
- Guan W.-J., Ni Z.-y., Hu Y.u., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S.C., Du B., Li L.-J., Zeng G., Yuen K.-Y., Chen R.-C., Tang C.-l., Wang T., Chen P.-Y., Xiang J., Li S.-Y., Wang J.-L., Liang Z.-J., Peng Y.-X., Wei L.i., Liu Y., Hu Y.-H., Peng P., Wang J.-M., Liu J.-Y., Chen Z., Li G., Zheng Z.-J., Qiu S.-Q., Luo J., Ye C.-J., Zhu S.-Y., Zhong N.-S. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. - DOI - PMC - PubMed
-
- Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., Taylor J., Spicer K., Bardossy A.C., Oakley L.P., Tanwar S., Dyal J.W., Harney J., Chisty Z., Bell J.M., Methner M., Paul P., Carlson C.M., McLaughlin H.P., Thornburg N., Tong S., Tamin A., Tao Y., Uehara A., Harcourt J., Clark S., Brostrom-Smith C., Page L.C., Kay M., Lewis J., Montgomery P., Stone N.D., Clark T.A., Honein M.A., Duchin J.S., Jernigan J.A. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020;382(22):2081–2090. - PMC - PubMed
-
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

