The dual role of glioma exosomal microRNAs: glioma eliminates tumor suppressor miR-1298-5p via exosomes to promote immunosuppressive effects of MDSCs
- PMID: 35501306
- PMCID: PMC9061735
- DOI: 10.1038/s41419-022-04872-z
The dual role of glioma exosomal microRNAs: glioma eliminates tumor suppressor miR-1298-5p via exosomes to promote immunosuppressive effects of MDSCs
Abstract
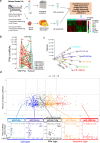
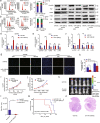
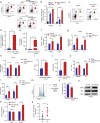
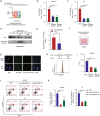
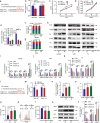
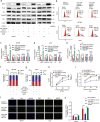
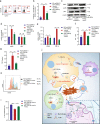
Clear evidence shows that tumors could secrete microRNAs (miRNAs) via exosomes to modulate the tumor microenvironment (TME). However, the mechanisms sorting specific miRNAs into exosomes are still unclear. In order to study the biological function and characterization of exosomal miRNAs, we performed whole-transcriptome sequencing in 59 patients' whole-course cerebrospinal fluid (CSF) small extracellular vesicles (sEV) and matched glioma tissue samples. The results demonstrate that miRNAs could be divided into exosome-enriched miRNAs (ExomiRNAs) and intracellular-retained miRNAs (CLmiRNAs), and exosome-enriched miRNAs generally play a dual role. Among them, miR-1298-5p was enriched in CSF exosomes and suppressed glioma progression in vitro and vivo experiments. Interestingly, exosomal miR-1298-5p could promote the immunosuppressive effects of myeloid-derived suppressor cells (MDSCs) to facilitate glioma. Therefore, we found miR-1298-5p had different effects on glioma cells and MDSCs. Mechanically, downstream signaling pathway analyses showed that miR-1298-5p plays distinct roles in glioma cells and MDSCs via targeting SETD7 and MSH2, respectively. Moreover, reverse verification was performed on the intracellular-retained miRNA miR-9-5p. Thus, we confirmed that tumor-suppressive miRNAs in glioma cells could be eliminated through exosomes and target tumor-associated immune cells to induce tumor-promoting phenotypes. Glioma could get double benefit from it. These findings uncover the mechanisms that glioma selectively sorts miRNAs into exosomes and modulates tumor immunity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Exosomal miR-1246 from glioma patient body fluids drives the differentiation and activation of myeloid-derived suppressor cells.Mol Ther. 2021 Dec 1;29(12):3449-3464. doi: 10.1016/j.ymthe.2021.06.023. Epub 2021 Jul 2. Mol Ther. 2021. PMID: 34217892 Free PMC article.
-
Tumor-derived exosomes deliver the tumor suppressor miR-3591-3p to induce M2 macrophage polarization and promote glioma progression.Oncogene. 2022 Oct;41(41):4618-4632. doi: 10.1038/s41388-022-02457-w. Epub 2022 Sep 9. Oncogene. 2022. PMID: 36085418 Free PMC article.
-
Glioma exosomes mediate the expansion and function of myeloid-derived suppressor cells through microRNA-29a/Hbp1 and microRNA-92a/Prkar1a pathways.Int J Cancer. 2019 Jun 15;144(12):3111-3126. doi: 10.1002/ijc.32052. Epub 2019 Jan 12. Int J Cancer. 2019. PMID: 30536597
-
The immunomodulatory role of exosomal microRNA networks in the crosstalk between tumor-associated myeloid-derived suppressor cells and tumor cells.Int Immunopharmacol. 2023 Jul;120:110267. doi: 10.1016/j.intimp.2023.110267. Epub 2023 Jun 3. Int Immunopharmacol. 2023. PMID: 37276829 Review.
-
Interaction Between microRNAs and Myeloid-Derived Suppressor Cells in Tumor Microenvironment.Front Immunol. 2022 May 11;13:883683. doi: 10.3389/fimmu.2022.883683. eCollection 2022. Front Immunol. 2022. PMID: 35634311 Free PMC article. Review.
Cited by
-
Exosomes-mediated crosstalk between glioma and immune cells in the tumor microenvironment.CNS Neurosci Ther. 2023 Aug;29(8):2074-2085. doi: 10.1111/cns.14239. Epub 2023 May 11. CNS Neurosci Ther. 2023. PMID: 37170647 Free PMC article. Review.
-
Constructing a signature based on the SIRT family to help the prognosis and treatment sensitivity in glioma patients.Front Genet. 2022 Dec 9;13:1035368. doi: 10.3389/fgene.2022.1035368. eCollection 2022. Front Genet. 2022. PMID: 36568393 Free PMC article.
-
Beyond the Bubble: A Debate on microRNA Sorting Into Extracellular Vesicles.Lab Invest. 2025 Feb;105(2):102206. doi: 10.1016/j.labinv.2024.102206. Epub 2024 Dec 6. Lab Invest. 2025. PMID: 39647608 Review.
-
Mechanisms Controlling MicroRNA Expression in Tumor.Cells. 2022 Sep 13;11(18):2852. doi: 10.3390/cells11182852. Cells. 2022. PMID: 36139427 Free PMC article. Review.
-
Exploring the roles and clinical potential of exosome-derived non-coding RNAs in glioma.IBRO Neurosci Rep. 2025 Feb 5;18:323-337. doi: 10.1016/j.ibneur.2025.01.015. eCollection 2025 Jun. IBRO Neurosci Rep. 2025. PMID: 40034544 Free PMC article. Review.
References
-
- Galon J, Bruni D. Tumor immunology and tumor evolution: intertwined histories. Immunity. 2020;52:55–81. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

