Bacterial communities associated with silage of different forage crops in Malaysian climate analysed using 16S amplicon metagenomics
- PMID: 35501317
- PMCID: PMC9061801
- DOI: 10.1038/s41598-022-08819-4
Bacterial communities associated with silage of different forage crops in Malaysian climate analysed using 16S amplicon metagenomics
Abstract
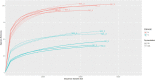
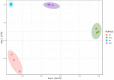
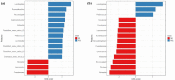
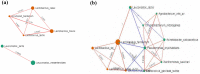
Silage produced in tropical countries is prone to spoilage because of high humidity and temperature. Therefore, determining indigenous bacteria as potential inoculants is important to improve silage quality. This study aimed to determine bacterial community and functional changes associated with ensiling using amplicon metagenomics and to predict potential bacterial additives associated with silage quality in the Malaysian climate. Silages of two forage crops (sweet corn and Napier) were prepared, and their fermentation properties and functional bacterial communities were analysed. After ensiling, both silages were predominated by lactic acid bacteria (LAB), and they exhibited good silage quality with significant increment in lactic acid, reductions in pH and water-soluble carbohydrates, low level of acetic acid and the absence of propionic and butyric acid. LAB consortia consisting of homolactic and heterolactic species were proposed to be the potential bacterial additives for sweet corn and Napier silage fermentation. Tax4fun functional prediction revealed metabolic pathways related to fermentation activities (bacterial division, carbohydrate transport and catabolism, and secondary metabolite production) were enriched in ensiled crops (p < 0.05). These results might suggest active transport and metabolism of plant carbohydrates into a usable form to sustain bacterial reproduction during silage fermentation, yielding metabolic products such as lactic acid. This research has provided a comprehensive understanding of bacterial communities before and after ensiling, which can be useful for desirable silage fermentation in Malaysia.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




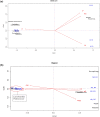
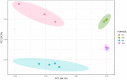

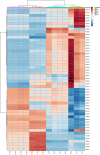

Similar articles
-
Impact of Saccharomyces cerevisiae and Lactobacillus buchneri on microbial communities during ensiling and aerobic spoilage of corn silage1.J Anim Sci. 2019 Mar 1;97(3):1273-1285. doi: 10.1093/jas/skz021. J Anim Sci. 2019. PMID: 30715344 Free PMC article.
-
Bacterial and fungal core microbiomes associated with small grain silages during ensiling and aerobic spoilage.BMC Microbiol. 2017 Mar 3;17(1):50. doi: 10.1186/s12866-017-0947-0. BMC Microbiol. 2017. PMID: 28253864 Free PMC article.
-
Effects of inoculants Lactobacillus brevis and Lactobacillus parafarraginis on the fermentation characteristics and microbial communities of corn stover silage.Sci Rep. 2017 Oct 19;7(1):13614. doi: 10.1038/s41598-017-14052-1. Sci Rep. 2017. PMID: 29051616 Free PMC article.
-
Silage review: Recent advances and future uses of silage additives.J Dairy Sci. 2018 May;101(5):3980-4000. doi: 10.3168/jds.2017-13839. J Dairy Sci. 2018. PMID: 29685273 Review.
-
The performance of lactic acid bacteria in silage production: A review of modern biotechnology for silage improvement.Microbiol Res. 2023 Jan;266:127212. doi: 10.1016/j.micres.2022.127212. Epub 2022 Sep 30. Microbiol Res. 2023. PMID: 36240665 Review.
Cited by
-
Effects of lactic acid bacteria inoculants on the nutrient composition, fermentation quality, and microbial diversity of whole-plant soybean-corn mixed silage.Front Microbiol. 2024 Apr 15;15:1347293. doi: 10.3389/fmicb.2024.1347293. eCollection 2024. Front Microbiol. 2024. PMID: 38686105 Free PMC article.
-
Utilisation of Lactiplantibacillus plantarum and propionic acid to improve silage quality of amaranth before and after wilting: fermentation quality, microbial communities, and their metabolic pathway.Front Microbiol. 2024 Jun 6;15:1415290. doi: 10.3389/fmicb.2024.1415290. eCollection 2024. Front Microbiol. 2024. PMID: 38903783 Free PMC article.
-
Effects of Moisture Content Gradient on Alfalfa Silage Quality, Odor, and Bacterial Community Revealed by Electronic Nose and GC-MS.Microorganisms. 2025 Feb 9;13(2):381. doi: 10.3390/microorganisms13020381. Microorganisms. 2025. PMID: 40005747 Free PMC article.
-
Analysis of the formation mechanism and regulation pathway of oat silage off-flavor based on microbial-metabolite interaction network.BMC Microbiol. 2025 Jul 30;25(1):464. doi: 10.1186/s12866-025-04166-2. BMC Microbiol. 2025. PMID: 40739180 Free PMC article.
-
Influences of Growth Stage and Ensiling Time on Fermentation Characteristics, Nitrite, and Bacterial Communities during Ensiling of Alfalfa.Plants (Basel). 2023 Dec 27;13(1):84. doi: 10.3390/plants13010084. Plants (Basel). 2023. PMID: 38202392 Free PMC article.
References
-
- Department of Veterinary Services Malaysia . Perangkaan Ternakan Livestock Statistics. Department of Veterinary Services Malaysia; 2021.
-
- Halim RA, Shampazurini S, Idris AB. Yield and nutritive quality of nine Napier grass varieties in Malaysia. Malays. J. Anim. Sci. 2013;16:37–44.
-
- Ortega-Gãmez R, et al. Nutritive quality of ten grasses during the rainy season in a hot-humid climate and ultisol soil. Trop. Subtrop. Agroecosyst. 2011;13:481.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

