The 4D Nucleome Data Portal as a resource for searching and visualizing curated nucleomics data
- PMID: 35501320
- PMCID: PMC9061818
- DOI: 10.1038/s41467-022-29697-4
The 4D Nucleome Data Portal as a resource for searching and visualizing curated nucleomics data
Erratum in
-
Author Correction: The 4D Nucleome Data Portal as a resource for searching and visualizing curated nucleomics data.Nat Commun. 2022 Nov 2;13(1):6561. doi: 10.1038/s41467-022-34458-4. Nat Commun. 2022. PMID: 36323678 Free PMC article. No abstract available.
Abstract
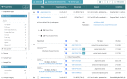
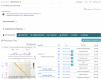
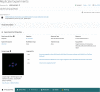
The 4D Nucleome (4DN) Network aims to elucidate the complex structure and organization of chromosomes in the nucleus and the impact of their disruption in disease biology. We present the 4DN Data Portal ( https://data.4dnucleome.org/ ), a repository for datasets generated in the 4DN network and relevant external datasets. Datasets were generated with a wide range of experiments, including chromosome conformation capture assays such as Hi-C and other innovative sequencing and microscopy-based assays probing chromosome architecture. All together, the 4DN data portal hosts more than 1800 experiment sets and 36000 files. Results of sequencing-based assays from different laboratories are uniformly processed and quality-controlled. The portal interface allows easy browsing, filtering, and bulk downloads, and the integrated HiGlass genome browser allows interactive visualization and comparison of multiple datasets. The 4DN data portal represents a primary resource for chromosome contact and other nuclear architecture data for the scientific community.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

