Transcription Factor 4 loss-of-function is associated with deficits in progenitor proliferation and cortical neuron content
- PMID: 35501322
- PMCID: PMC9061776
- DOI: 10.1038/s41467-022-29942-w
Transcription Factor 4 loss-of-function is associated with deficits in progenitor proliferation and cortical neuron content
Erratum in
-
Author Correction: Transcription Factor 4 loss-of-function is associated with deficits in progenitor proliferation and cortical neuron content.Nat Commun. 2025 Feb 12;16(1):1565. doi: 10.1038/s41467-025-56808-8. Nat Commun. 2025. PMID: 39939331 Free PMC article. No abstract available.
Abstract
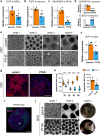
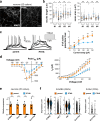
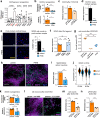
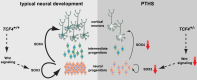
Transcription Factor 4 (TCF4) has been associated with autism, schizophrenia, and other neuropsychiatric disorders. However, how pathological TCF4 mutations affect the human neural tissue is poorly understood. Here, we derive neural progenitor cells, neurons, and brain organoids from skin fibroblasts obtained from children with Pitt-Hopkins Syndrome carrying clinically relevant mutations in TCF4. We show that neural progenitors bearing these mutations have reduced proliferation and impaired capacity to differentiate into neurons. We identify a mechanism through which TCF4 loss-of-function leads to decreased Wnt signaling and then to diminished expression of SOX genes, culminating in reduced progenitor proliferation in vitro. Moreover, we show reduced cortical neuron content and impaired electrical activity in the patient-derived organoids, phenotypes that were rescued after correction of TCF4 expression or by pharmacological modulation of Wnt signaling. This work delineates pathological mechanisms in neural cells harboring TCF4 mutations and provides a potential target for therapeutic strategies for genetic disorders associated with this gene.
© 2022. The Author(s).
Conflict of interest statement
Dr. Muotri is the co-founder of and has an equity interest in TISMOO, a company dedicated to genetic analysis and human brain organogenesis, focusing on therapeutic applications customized to autism spectrum disorders and other neurological diseases. The terms of this arrangement have been reviewed and approved by the University of California San Diego, in accordance with its conflict-of-interest policies. UCSD has filed a patent application (WO2022072709A1), in which F.P. and A.R.M. are inventors, containing some results regarding the TCF4 correction overexpression strategy described in this paper. The patent was published on 04-07-2022. Other authors declare no competing interests.
Figures






References
-
- de Pontual, L. et al. Mutational, functional, and expression studies of the TCF4 gene in Pitt-Hopkins syndrome. Hum. Mutat.30, 669–676 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

