Functional and pathologic association of aminoacyl-tRNA synthetases with cancer
- PMID: 35501376
- PMCID: PMC9166799
- DOI: 10.1038/s12276-022-00765-5
Functional and pathologic association of aminoacyl-tRNA synthetases with cancer
Abstract
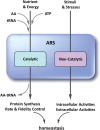
Although key tumorigenic and tumor-suppressive factors have been unveiled over the last several decades, cancer remains the most life-threatening disease. Multiomic analyses of patient samples and an in-depth understanding of tumorigenic processes have rapidly revealed unexpected pathologic associations of new cellular factors previously overlooked in cancer biology. In this regard, the newly discovered activities of human aminoacyl-tRNA synthases (ARSs) deserve attention not only for their pathological significance in tumorigenesis but also regarding diagnostic and therapeutic implications. ARSs are not only essential enzymes covalently linking substrate amino acids to cognate tRNAs for protein synthesis but also function as regulators of cellular processes by sensing different cellular conditions. With their catalytic role in protein synthesis and their regulatory role in homeostasis, functional alterations or dysregulation of ARSs might be pathologically associated with tumorigenesis. This review focuses on the potential implications of ARS genes and proteins in different aspects of cancer based on various bioinformatic analyses and experimental data. We also review their diverse activities involving extracellular secretion, protein-protein interactions, and amino acid sensing, which are related to cancers. The newly discovered cancer-related activities of ARSs are expected to provide new opportunities for detecting, preventing and curing cancers.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

