Plasma proteome analyses in individuals of European and African ancestry identify cis-pQTLs and models for proteome-wide association studies
- PMID: 35501419
- PMCID: PMC9236177
- DOI: 10.1038/s41588-022-01051-w
Plasma proteome analyses in individuals of European and African ancestry identify cis-pQTLs and models for proteome-wide association studies
Abstract
Improved understanding of genetic regulation of the proteome can facilitate identification of the causal mechanisms for complex traits. We analyzed data on 4,657 plasma proteins from 7,213 European American (EA) and 1,871 African American (AA) individuals from the Atherosclerosis Risk in Communities study, and further replicated findings on 467 AA individuals from the African American Study of Kidney Disease and Hypertension study. Here, we identified 2,004 proteins in EA and 1,618 in AA, with most overlapping, which showed associations with common variants in cis-regions. Availability of AA samples led to smaller credible sets and notable number of population-specific cis-protein quantitative trait loci. Elastic Net produced powerful models for protein prediction in both populations. An application of proteome-wide association studies to serum urate and gout implicated several proteins, including IL1RN, revealing the promise of the drug anakinra to treat acute gout flares. Our study demonstrates the value of large and diverse ancestry study to investigate the genetic mechanisms of molecular phenotypes and their relationship with complex traits.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests Statement
Proteomic assays in ARIC were conducted free of charge as part of a data exchange agreement with Soma Logic. The authors declare no other competing interests.
Figures
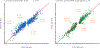


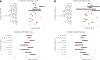

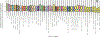
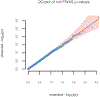






References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG010480/HG/NHGRI NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- R01 AR073178/AR/NIAMS NIH HHS/United States
- HHSN268201700002C/HL/NHLBI NIH HHS/United States
- HHSN268201700001I/HL/NHLBI NIH HHS/United States
- HHSN268201700004I/HL/NHLBI NIH HHS/United States
- R01 HL148218/HL/NHLBI NIH HHS/United States
- HHSN268201700004C/HL/NHLBI NIH HHS/United States
- R01 HL134320/HL/NHLBI NIH HHS/United States
- HHSN268201700003I/HL/NHLBI NIH HHS/United States
- R01 DK124399/DK/NIDDK NIH HHS/United States
- HHSN268201700005C/HL/NHLBI NIH HHS/United States
- HHSN268201700001C/HL/NHLBI NIH HHS/United States
- HHSN268201700003C/HL/NHLBI NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- HHSN268201700002I/HL/NHLBI NIH HHS/United States
- HHSN268201700005I/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

