Characterization of cell-cell communication in autistic brains with single-cell transcriptomes
- PMID: 35501678
- PMCID: PMC9059394
- DOI: 10.1186/s11689-022-09441-1
Characterization of cell-cell communication in autistic brains with single-cell transcriptomes
Abstract
Background: Autism spectrum disorder is a neurodevelopmental disorder, affecting 1-2% of children. Studies have revealed genetic and cellular abnormalities in the brains of affected individuals, leading to both regional and distal cell communication deficits.
Methods: Recent application of single-cell technologies, especially single-cell transcriptomics, has significantly expanded our understanding of brain cell heterogeneity and further demonstrated that multiple cell types and brain layers or regions are perturbed in autism. The underlying high-dimensional single-cell data provides opportunities for multilevel computational analysis that collectively can better deconvolute the molecular and cellular events altered in autism. Here, we apply advanced computation and pattern recognition approaches on single-cell RNA-seq data to infer and compare inter-cell-type signaling communications in autism brains and controls.
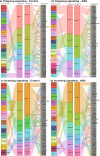
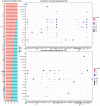
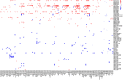
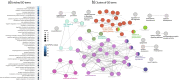
Results: Our results indicate that at a global level, there are cell-cell communication differences in autism in comparison with controls, largely involving neurons as both signaling senders and receivers, but glia also contribute to the communication disruption. Although the magnitude of changes is moderate, we find that excitatory and inhibitor neurons are involved in multiple intercellular signaling that exhibits increased strengths in autism, such as NRXN and CNTN signaling. Not all genes in the intercellular signaling pathways show differential expression, but genes in the affected pathways are enriched for axon guidance, synapse organization, neuron migration, and other critical cellular functions. Furthermore, those genes are highly connected to and enriched for genes previously associated with autism risks.
Conclusions: Overall, our proof-of-principle computational study using single-cell data uncovers key intercellular signaling pathways that are potentially disrupted in the autism brains, suggesting that more studies examining cross-cell type effects can be valuable for understanding autism pathogenesis.
Keywords: Autism; Brain; Cell-cell communication; Ligand-receptor; Network; Single-cell RNA-seq.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- American Psychiatric Association . Highlights of changes from DSM-IV-TR to DSM-5. 2013.
-
- Maenner MJ, Shaw KA, Baio J, Washington A, Patrick M, DiRienzo M, et al. Prevalence of autism spectrum disorder among children aged 8 years-autism and developmental disabilities monitoring network, 11 sites, United States, 2016. MMWR Surveill Summ. 2020;69:1–12. doi: 10.15585/mmwr.ss6904a1. - DOI - PMC - PubMed

