A large-scale population based organelle pan-genomes construction and phylogeny analysis reveal the genetic diversity and the evolutionary origins of chloroplast and mitochondrion in Brassica napus L
- PMID: 35501686
- PMCID: PMC9063048
- DOI: 10.1186/s12864-022-08573-x
A large-scale population based organelle pan-genomes construction and phylogeny analysis reveal the genetic diversity and the evolutionary origins of chloroplast and mitochondrion in Brassica napus L
Erratum in
-
Correction: A large-scale population based organelle pan-genomes construction and phylogeny analysis reveal the genetic diversity and the evolutionary origins of chloroplast and mitochondrion in Brassica napus L.BMC Genomics. 2023 Nov 27;24(1):716. doi: 10.1186/s12864-023-09812-5. BMC Genomics. 2023. PMID: 38012560 Free PMC article. No abstract available.
Abstract
Background: Allotetraploid oilseed rape (Brassica napus L.) is an important worldwide oil-producing crop. The origin of rapeseed is still undetermined due to the lack of wild resources. Despite certain genetic architecture and phylogenetic studies have been done focus on large group of Brassica nuclear genomes, the organelle genomes information under global pattern is largely unknown, which provide unique material for phylogenetic studies of B. napus. Here, based on de novo assemblies of 1,579 B. napus accessions collected globally, we constructed the chloroplast and mitochondrial pan-genomes of B. napus, and investigated the genetic diversity, phylogenetic relationships of B. napus, B. rapa and B. oleracea.
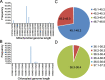
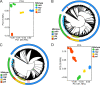
Results: Based on mitotype-specific markers and mitotype-variant ORFs, four main cytoplasmic haplotypes were identified in our groups corresponding the nap, pol, ole, and cam mitotypes, among which the structure of chloroplast genomes was more conserved without any rearrangement than mitochondrial genomes. A total of 2,092 variants were detected in chloroplast genomes, whereas only 326 in mitochondrial genomes, indicating that chloroplast genomes exhibited a higher level of single-base polymorphism than mitochondrial genomes. Based on whole-genome variants diversity analysis, eleven genetic difference regions among different cytoplasmic haplotypes were identified on chloroplast genomes. The phylogenetic tree incorporating accessions of the B. rapa, B. oleracea, natural and synthetic populations of B. napus revealed multiple origins of B. napus cytoplasm. The cam-type and pol-type were both derived from B. rapa, while the ole-type was originated from B. oleracea. Notably, the nap-type cytoplasm was identified in both the B. rapa population and the synthetic B. napus, suggesting that B. rapa might be the maternal ancestor of nap-type B. napus.
Conclusions: The phylogenetic results provide novel insights into the organelle genomic evolution of Brassica species. The natural rapeseeds contained at least four cytoplastic haplotypes, of which the predominant nap-type might be originated from B. rapa. Besides, the organelle pan-genomes and the overall variation data offered useful resources for analysis of cytoplasmic inheritance related agronomical important traits of rapeseed, which can substantially facilitate the cultivation and improvement of rapeseed varieties.
Keywords: Brassica; Cytoplasm haplotype; Maternal ancestor; Mitotype; Organelle pan-genome; Rapeseed.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
High-throughput multiplex cpDNA resequencing clarifies the genetic diversity and genetic relationships among Brassica napus, Brassica rapa and Brassica oleracea.Plant Biotechnol J. 2016 Jan;14(1):409-18. doi: 10.1111/pbi.12395. Epub 2015 Jun 1. Plant Biotechnol J. 2016. PMID: 26031705 Free PMC article.
-
Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers.BMC Plant Biol. 2010 Mar 29;10:54. doi: 10.1186/1471-2229-10-54. BMC Plant Biol. 2010. PMID: 20350303 Free PMC article.
-
Comparison of the cytoplastic genomes by resequencing: insights into the genetic diversity and the phylogeny of the agriculturally important genus Brassica.BMC Genomics. 2020 Jul 13;21(1):480. doi: 10.1186/s12864-020-06889-0. BMC Genomics. 2020. PMID: 32660507 Free PMC article.
-
Exploring the gene pool of Brassica napus by genomics-based approaches.Plant Biotechnol J. 2021 Sep;19(9):1693-1712. doi: 10.1111/pbi.13636. Epub 2021 Jun 17. Plant Biotechnol J. 2021. PMID: 34031989 Free PMC article. Review.
-
A review of sources of resistance to turnip yellows virus (TuYV) in Brassica species.Ann Appl Biol. 2023 Nov;183(3):200-208. doi: 10.1111/aab.12842. Epub 2023 Aug 15. Ann Appl Biol. 2023. PMID: 38515540 Free PMC article. Review.
Cited by
-
Maternal Donor and Genetic Variation of Lagerstroemia indica Cultivars.Int J Mol Sci. 2023 Feb 10;24(4):3606. doi: 10.3390/ijms24043606. Int J Mol Sci. 2023. PMID: 36835020 Free PMC article.
-
Assembly and analysis of stephania japonica mitochondrial genome provides new insights into its identification and energy metabolism.BMC Genomics. 2025 Feb 24;26(1):185. doi: 10.1186/s12864-025-11359-6. BMC Genomics. 2025. PMID: 39994544 Free PMC article.
-
Correction: A large-scale population based organelle pan-genomes construction and phylogeny analysis reveal the genetic diversity and the evolutionary origins of chloroplast and mitochondrion in Brassica napus L.BMC Genomics. 2023 Nov 27;24(1):716. doi: 10.1186/s12864-023-09812-5. BMC Genomics. 2023. PMID: 38012560 Free PMC article. No abstract available.
-
The complete chloroplast genome of Brassica rapa var. purpuraria (L.H.Bailey) Kitam 1950 and its phylogenetic analysis.Mitochondrial DNA B Resour. 2024 Jan 24;9(1):143-147. doi: 10.1080/23802359.2024.2305403. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 38274856 Free PMC article.
-
Chloroplast Genomes Evolution and Phylogenetic Relationships of Caragana species.Int J Mol Sci. 2024 Jun 20;25(12):6786. doi: 10.3390/ijms25126786. Int J Mol Sci. 2024. PMID: 38928490 Free PMC article.
References
-
- Nagaharu U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot. 1935;7:389–452.
-
- Schmidt R, van Bancroft I. Genetics and Genomics of the Brassicaceae. New York: Springer; 2011. pp. 585–596.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

