Comparison of an automated DNA extraction and 16S rDNA real time PCR/sequencing diagnostic method using optimized reagents with culture during a 15-month study using specimens from sterile body sites
- PMID: 35501697
- PMCID: PMC9063205
- DOI: 10.1186/s12866-022-02542-w
Comparison of an automated DNA extraction and 16S rDNA real time PCR/sequencing diagnostic method using optimized reagents with culture during a 15-month study using specimens from sterile body sites
Abstract
Background: 16S rDNA-PCR for the identification of a bacterial species is an established method. However, the DNA extraction reagents as well as the PCR reagents may contain residual bacterial DNA, which consequently generates false-positive PCR results. Additionally, previously used methods are frequently time-consuming. Here, we describe the results obtained with a new technology that uses DNA-free reagents for automated DNA extraction and subsequent real time PCR using sterile clinical specimens.
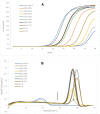
Results: In total, we compared 803 clinical specimens using real time PCR and culturing. The clinical specimens were mainly of orthopedic origin received at our diagnostic laboratory. In 595 (74.1%) samples, the results were concordant negative, and in 102 (12.7%) the results were concordant positive. A total of 170 (21.2%) clinical specimens were PCR-positive, of which 62 (36.5% from PCR positive, 7.7% in total) gave an additional benefit to the patient since only the PCR result was positive. Many of these 62 positive specimens were strongly positive based on crossingpoint values (54% < Cp 30), and these 62 positive clinical specimens were diagnosed as medically relevant as well. Thirty-eight (4.2%) clinical specimens were culture-positive (25 of them were only enrichment culture positive) but PCR-negative, mainly for S. epidermidis, S. aureus and C. acnes. The turnaround times for negative specimens were 4 hours (automated DNA extraction and real time PCR) and 1 working day for positive specimens (including Sanger sequencing). Melting-curve analysis of SYBR Green-PCR enables the differentiation of specific and unspecific PCR products. Using Ripseq, even mixed infections of 2 bacterial species could be resolved.
Conclusions: For endocarditis cases, the added benefit of PCR is obvious. The crucial innovations of the technology enable timely reporting of explicit reliable results for adequate treatment of patients. Clinical specimens with truly PCR-positive but culture-negative results represent an additional benefit for patients. Very few results at the detection limit still have to be critically examined.
Keywords: 16S rDNA real time PCR; Additional benefit; DNA-free reagents; Evolution of methods; Mixed sequences.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
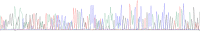
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

