Circulating lnc-LOC as a novel noninvasive biomarker in the treatment surveillance of acute promyelocytic leukaemia
- PMID: 35501730
- PMCID: PMC9059359
- DOI: 10.1186/s12885-022-09621-1
Circulating lnc-LOC as a novel noninvasive biomarker in the treatment surveillance of acute promyelocytic leukaemia
Abstract
Background: Acute promyelocytic leukaemia (APL) is a unique subtype of acute myeloid leukaemia (AML) characterized by haematopoietic failure caused by the accumulation of abnormal promyelocytic cells in bone marrow (BM). However, indispensable BM biopsy frequently afflicts patients in leukaemia surveillance, which increases the burden on patients and reduces compliance. This study aimed to explore whether the novel circulating long noncoding RNA LOC100506453 (lnc-LOC) could be a target in diagnosis, assess the treatment response and supervise the minimal residual disease (MRD) of APL, thereby blazing a trail in noninvasive lncRNA biomarkers of APL.
Methods: Our study comprised 100 patients (40 with APL and 60 with non-APL AML) and 60 healthy donors. BM and peripheral blood (PB) sample collection was accomplished from APL patients at diagnosis and postinduction. Quantitative real-time PCR (qRT-PCR) was conducted to evaluate lnc-LOC expression. A receiver operating characteristic (ROC) analysis was implemented to analyse the value of lnc-LOC in the diagnosis of APL and treatment monitoring. For statistical analysis, the Mann-Whitney U test, a t test, and Spearman's rank correlation test were utilized.
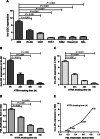
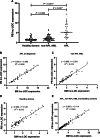
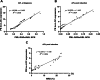
Results: Our results showed that BM lnc-LOC expression was significantly different between APL and healthy donors and non-APL AML. lnc-LOC was drastically downregulated in APL patients' BM after undergoing induction therapy. Lnc-LOC was upregulated in APL cell lines and downregulated after all-trans retinoic acid (ATRA)-induced myeloid differentiation, preliminarily verifying that lnc-LOC has the potential to be considered a treatment monitoring biomarker. PB lnc-LOC was positively correlated with BM lnc-LOC in APL patients, non-APL AML patients and healthy donors and decreased sharply after complete remission (CR). However, upregulated lnc-LOC was manifested in relapsed-refractory patients. A positive correlation was revealed between PB lnc-LOC and PML-RARα transcript levels in BM samples. Furthermore, we observed a positive correlation between PB lnc-LOC and BM lnc-LOC expression in APL patients, suggesting that lnc-LOC can be utilized as a noninvasive biomarker for MRD surveillance.
Conclusions: Our study demonstrated that PB lnc-LOC might serve as a novel noninvasive biomarker in the treatment surveillance of APL, and it innovated the investigation and application of newly found lncRNAs in APL noninvasive biomarkers used in diagnosis and detection.
Keywords: Acute promyelocytic leukaemia; Lnc-LOC; Minimal residual disease; Noninvasive biomarker; Surveillance.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
miR-638 in circulating leukaemia cells as a non-invasive biomarker in diagnosis, treatment response and MRD surveillance of acute promyelocytic leukaemia.Cancer Biomark. 2020;29(1):125-137. doi: 10.3233/CBM-190899. Cancer Biomark. 2020. PMID: 32568176
-
Genome‑wide profiling of lncRNA expression patterns in patients with acute promyelocytic leukemia with differentiation therapy.Oncol Rep. 2018 Sep;40(3):1601-1613. doi: 10.3892/or.2018.6521. Epub 2018 Jun 25. Oncol Rep. 2018. PMID: 29956795
-
Transcription therapy for acute promyelocytic leukaemia.Expert Opin Investig Drugs. 2000 Feb;9(2):329-46. doi: 10.1517/13543784.9.2.329. Expert Opin Investig Drugs. 2000. PMID: 11060680 Review.
-
Retinoic Acid-Induced Gene G(RIG-G) as a Novel Monitoring Biomarker in Leukemia and Its Clinical Applications.Genes (Basel). 2021 Jul 2;12(7):1035. doi: 10.3390/genes12071035. Genes (Basel). 2021. PMID: 34356051 Free PMC article.
-
Treatment of acute promyelocytic leukaemia with all-trans retinoic acid and arsenic trioxide: a paradigm of synergistic molecular targeting therapy.Philos Trans R Soc Lond B Biol Sci. 2007 Jun 29;362(1482):959-71. doi: 10.1098/rstb.2007.2026. Philos Trans R Soc Lond B Biol Sci. 2007. PMID: 17317642 Free PMC article. Review.
Cited by
-
Cuproptosis-related lncRNA signature for prognostic prediction in patients with acute myeloid leukemia.BMC Bioinformatics. 2023 Feb 3;24(1):37. doi: 10.1186/s12859-023-05148-9. BMC Bioinformatics. 2023. PMID: 36737692 Free PMC article.
-
Exploration of the Regulatory Network of Programmed Cell Death Genes in Rheumatoid Arthritis Based on Blood-Derived circRNA Transcriptome Information and Single-Cell Multi-omics Data.Biochem Genet. 2024 Dec 10. doi: 10.1007/s10528-024-10989-x. Online ahead of print. Biochem Genet. 2024. PMID: 39656402
-
Hsa_circ_0006010 and hsa_circ_0002903 in peripheral blood serve as novel diagnostic, surveillance and prognostic biomarkers for disease progression in chronic myeloid leukemia.BMC Cancer. 2024 Sep 20;24(1):1172. doi: 10.1186/s12885-024-12943-x. BMC Cancer. 2024. PMID: 39304860 Free PMC article.
References
-
- Gabert J, Beillard E, van der Velden VH, Bi W, Grimwade D, Pallisgaard N, et al. Standardization and quality control studies of 'real-time' quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia - a Europe against Cancer program. Leukemia. 2003;17(12):2318–2357. doi: 10.1038/sj.leu.2403135. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- LGF20H200005/The Basic Public Welfare Technology Research Project of Zhejiang Province
- 2021KY216/The Medical and Health Research Science and Technology Plan Project of Zhejiang Province
- Y20190090/The Basic Scientific Research Project of Wenzhou City
- 18331203/The Lin He's New Medicine and Clinical Translation Academician Workstation Research Fund
LinkOut - more resources
Full Text Sources
Medical

