A systematic analysis of gene-gene interaction in multiple sclerosis
- PMID: 35501860
- PMCID: PMC9063218
- DOI: 10.1186/s12920-022-01247-3
A systematic analysis of gene-gene interaction in multiple sclerosis
Abstract
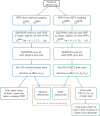
Background: For the most part, genome-wide association studies (GWAS) have only partially explained the heritability of complex diseases. One of their limitations is to assume independent contributions of individual variants to the phenotype. Many tools have therefore been developed to investigate the interactions between distant loci, or epistasis. Among them, the recently proposed EpiGWAS models the interactions between a target variant and the rest of the genome. However, applying this approach to studying interactions along all genes of a disease map is not straightforward. Here, we propose a pipeline to that effect, which we illustrate by investigating a multiple sclerosis GWAS dataset from the Wellcome Trust Case Control Consortium 2 through 19 disease maps from the MetaCore pathway database.
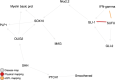
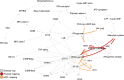
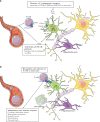
Results: For each disease map, we build an epistatic network by connecting the genes that are deemed to interact. These networks tend to be connected, complementary to the disease maps and contain hubs. In addition, we report 4 epistatic gene pairs involving missense variants, and 25 gene pairs with a deleterious epistatic effect mediated by eQTLs. Among these, we highlight the interaction of GLI-1 and SUFU, and of IP10 and NF-[Formula: see text]B, as they both match known biological interactions. The latter pair is particularly promising for therapeutic development, as both genes have known inhibitors.
Conclusions: Our study showcases the ability of EpiGWAS to uncover biologically interpretable epistatic interactions that are potentially actionable for the development of combination therapy.
Keywords: Causal inference; Epistasis; GWAS; Gene–gene interaction; Multiple sclerosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Gene, pathway and network frameworks to identify epistatic interactions of single nucleotide polymorphisms derived from GWAS data.BMC Syst Biol. 2012;6 Suppl 3(Suppl 3):S15. doi: 10.1186/1752-0509-6-S3-S15. Epub 2012 Dec 17. BMC Syst Biol. 2012. PMID: 23281810 Free PMC article.
-
Epistatic Gene-Based Interaction Analyses for Glaucoma in eMERGE and NEIGHBOR Consortium.PLoS Genet. 2016 Sep 13;12(9):e1006186. doi: 10.1371/journal.pgen.1006186. eCollection 2016 Sep. PLoS Genet. 2016. PMID: 27623284 Free PMC article.
-
A Genome-Wide Association Analysis Reveals Epistatic Cancellation of Additive Genetic Variance for Root Length in Arabidopsis thaliana.PLoS Genet. 2015 Sep 23;11(9):e1005541. doi: 10.1371/journal.pgen.1005541. eCollection 2015. PLoS Genet. 2015. PMID: 26397943 Free PMC article.
-
Post genome-wide association analysis: dissecting computational pathway/network-based approaches.Brief Bioinform. 2019 Mar 25;20(2):690-700. doi: 10.1093/bib/bby035. Brief Bioinform. 2019. PMID: 29701762 Free PMC article. Review.
-
The Genetics of Multiple Sclerosis: From 0 to 200 in 50 Years.Trends Genet. 2017 Dec;33(12):960-970. doi: 10.1016/j.tig.2017.09.004. Epub 2017 Oct 5. Trends Genet. 2017. PMID: 28987266 Free PMC article. Review.
Cited by
-
Heterogeneous network approaches to protein pathway prediction.Comput Struct Biotechnol J. 2024 Jun 27;23:2727-2739. doi: 10.1016/j.csbj.2024.06.022. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39035835 Free PMC article. Review.
-
MR-GGI: accurate inference of gene-gene interactions using Mendelian randomization.BMC Bioinformatics. 2024 May 15;25(1):192. doi: 10.1186/s12859-024-05808-4. BMC Bioinformatics. 2024. PMID: 38750431 Free PMC article.
-
Subtypes of relapsing-remitting multiple sclerosis identified by network analysis.Front Digit Health. 2023 Jan 11;4:1063264. doi: 10.3389/fdgth.2022.1063264. eCollection 2022. Front Digit Health. 2023. PMID: 36714613 Free PMC article.
-
Elevated genetic risk for multiple sclerosis emerged in steppe pastoralist populations.Nature. 2024 Jan;625(7994):321-328. doi: 10.1038/s41586-023-06618-z. Epub 2024 Jan 10. Nature. 2024. PMID: 38200296 Free PMC article.
-
Association of Epistatic Effects of lncRNA GAS5, miR-146a, IRAK-1, and miR-155 Genetic Variants with Multiple Sclerosis Risk and Severity.Mol Neurobiol. 2025 Aug;62(8):10742-10764. doi: 10.1007/s12035-025-04876-8. Epub 2025 Apr 15. Mol Neurobiol. 2025. PMID: 40234289 Free PMC article.
References
-
- Shaffer JP. Multiple hypothesis testing. Annu Rev Psychol. 1995;46(1):561–584. doi: 10.1146/annurev.ps.46.020195.003021. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

