Salvianolic Acid B Suppresses Non-Small-Cell Lung Cancer Metastasis through PKM2-Independent Metabolic Reprogramming
- PMID: 35502178
- PMCID: PMC9056207
- DOI: 10.1155/2022/9302403
Salvianolic Acid B Suppresses Non-Small-Cell Lung Cancer Metastasis through PKM2-Independent Metabolic Reprogramming
Abstract
Objective: Salvianolic acid B (Sal B) has been demonstrated to be a potential chemoprevention agent for several cancers. Herein, we investigated the pharmacological function of Sal B on non-small-cell lung cancer (NSCLC) metastasis.
Methods: Two NSCLC cell lines (NCI-H2030 and NCI-H1650) were disposed of by 200 μM Sal B or 10 μM PKM2 agonist TEPP-46. Wound healing and transwell experiments were implemented for analyzing migratory and invasive capacities. Epithelial-to-mesenchymal transition (EMT) markers β-catenin and E-cadherin were measured via western blotting. Cellular bioenergetics were evaluated with glucose uptake, lactate production, enolase activity, cellular ATP levels, as well as seahorse-based oxygen consumption rate (OCR), extracellular acidification rate (ECAR) analysis. Metabolic reprogramming markers PKM2, LDHA, and GLUT1 were detected via western blotting and immunofluorescence.
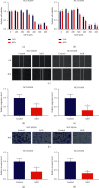
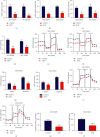
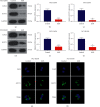
Results: The results showed that Sal B disposal weakened the migration and invasion of NCI-H2030 and NCI-H1650 cells and inactivated the EMT process according to downregulation of β-catenin and upregulation of E-cadherin. Sal B-treated NSCLC cells displayed decreased glucose uptake, lactate production, enolase activity, cellular ATP levels, OCR, and ECAR, indicating a reduction in metabolic reprogramming. Additionally, Sal B downregulated the expression of PKM2, LDHA, and GLUT1. TEPP-46 may reverse the inhibitory effect of Sal B on metastasis as well as metabolic reprogramming.
Conclusion: Our findings provide evidence that Sal B enables to weaken NSCLC metastasis through PKM2-independent metabolic reprogramming, which sheds light on the promising therapeutic usage of Sal B in treating NSCLC.
Copyright © 2022 Hong Zhang et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Salvianolic acid B acts against non‑small cell lung cancer A549 cells via inactivation of the MAPK and Smad2/3 signaling pathways.Mol Med Rep. 2022 May;25(5):184. doi: 10.3892/mmr.2022.12700. Epub 2022 Mar 29. Mol Med Rep. 2022. PMID: 35348194 Free PMC article.
-
Oxymatrine Inhibits Colorectal Cancer Metastasis via Attenuating PKM2-Mediated Aerobic Glycolysis.Cancer Manag Res. 2020 Oct 1;12:9503-9513. doi: 10.2147/CMAR.S267686. eCollection 2020. Cancer Manag Res. 2020. PMID: 33061637 Free PMC article.
-
[Effect of salvianolic acid B on high-glucose induced renal tubular epithelial-mesenchymal transition in rats and its mechanism].Zhongguo Zhong Yao Za Zhi. 2020 Aug;45(16):3922-3930. doi: 10.19540/j.cnki.cjcmm.20200405.403. Zhongguo Zhong Yao Za Zhi. 2020. PMID: 32893590 Chinese.
-
Salinomycin suppresses TGF-β1-induced EMT by down-regulating MMP-2 and MMP-9 via the AMPK/SIRT1 pathway in non-small cell lung cancer.Int J Med Sci. 2021 Jan 1;18(3):715-726. doi: 10.7150/ijms.50080. eCollection 2021. Int J Med Sci. 2021. PMID: 33437206 Free PMC article.
-
Salvianolic acid B suppresses EMT and apoptosis to lessen drug resistance through AKT/mTOR in gastric cancer cells.Cytotechnology. 2021 Feb;73(1):49-61. doi: 10.1007/s10616-020-00441-4. Epub 2020 Nov 17. Cytotechnology. 2021. PMID: 33505113 Free PMC article.
Cited by
-
Salvianolic Acid B: A Review of Pharmacological Effects, Safety, Combination Therapy, New Dosage Forms, and Novel Drug Delivery Routes.Pharmaceutics. 2023 Aug 29;15(9):2235. doi: 10.3390/pharmaceutics15092235. Pharmaceutics. 2023. PMID: 37765204 Free PMC article. Review.
-
Salvianolic Acid B Significantly Suppresses the Migration of Melanoma Cells via Direct Interaction with β-Actin.Molecules. 2024 Feb 19;29(4):906. doi: 10.3390/molecules29040906. Molecules. 2024. PMID: 38398656 Free PMC article.
-
Effect of Thymbra capitata (L.) Cav. on Inflammation, Senescence and Cell Migration.Nutrients. 2023 Apr 17;15(8):1930. doi: 10.3390/nu15081930. Nutrients. 2023. PMID: 37111149 Free PMC article.
-
Salvianolic acid B from Salvia miltiorrhiza bunge: A potential antitumor agent.Front Pharmacol. 2022 Oct 25;13:1042745. doi: 10.3389/fphar.2022.1042745. eCollection 2022. Front Pharmacol. 2022. PMID: 36386172 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

