Genome-Wide Knockout Screen Identifies Human Sialomucin CD164 as an Essential Entry Factor for Lymphocytic Choriomeningitis Virus
- PMID: 35502904
- PMCID: PMC9239079
- DOI: 10.1128/mbio.00205-22
Genome-Wide Knockout Screen Identifies Human Sialomucin CD164 as an Essential Entry Factor for Lymphocytic Choriomeningitis Virus
Abstract
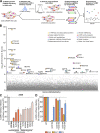
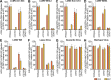
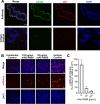
Lymphocytic choriomeningitis virus (LCMV) is a well-studied mammarenavirus that can be fatal in congenital infections. However, our understanding of LCMV and its interactions with human host factors remains incomplete. Here, host determinants affecting LCMV infection were investigated through a genome-wide CRISPR knockout screen in A549 cells, a human lung adenocarcinoma line. We identified and validated a variety of novel host factors that play a functional role in LCMV infection. Among these, knockout of the sialomucin CD164, a heavily glycosylated transmembrane protein, was found to ablate infection with multiple LCMV strains but not other hemorrhagic mammarenaviruses in several cell types. Further characterization revealed a dependency of LCMV entry on the cysteine-rich domain of CD164, including an N-linked glycosylation site at residue 104 in that region. Given the documented role of LCMV with respect to transplacental human infections, CD164 expression was investigated in human placental tissue and placental cell lines. CD164 was found to be highly expressed in the cytotrophoblast cells, an initial contact site for pathogens within the placenta, and LCMV infection in placental cells was effectively blocked using a monoclonal antibody specific to the cysteine-rich domain of CD164. Together, this study identifies novel factors associated with LCMV infection of human tissues and highlights the importance of CD164, a sialomucin that previously had not been associated with viral infection. IMPORTANCE Lymphocytic choriomeningitis virus (LCMV) is a human-pathogenic mammarenavirus that can be fatal in congenital infections. Although frequently used in the study of persistent infections in the field of immunology, aspects of this virus's life cycle remain incomplete. For example, while viral entry has been shown to depend on a cell adhesion molecule, DAG1, genetic knockout of this gene allows for residual viral infection, implying that additional receptors can mediate cell entry. The significance of our study is the identification of host factors important for successful infection, including the sialomucin CD164, which had not been previously associated with viral infection. We demonstrated that CD164 is essential for LCMV entry into human cells and can serve as a possible therapeutic target for treatment of congenital infection.
Keywords: CD164; CRISPR screen; lymphocytic choriomeningitis virus; viral entry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
CD164 is a host factor for lymphocytic choriomeningitis virus entry.Proc Natl Acad Sci U S A. 2022 Mar 8;119(10):e2119676119. doi: 10.1073/pnas.2119676119. Epub 2022 Mar 2. Proc Natl Acad Sci U S A. 2022. PMID: 35235462 Free PMC article.
-
Cellular endosomal potassium ion flux regulates arenavirus uncoating during virus entry.mBio. 2024 Jul 17;15(7):e0168423. doi: 10.1128/mbio.01684-23. Epub 2024 Jun 14. mBio. 2024. PMID: 38874413 Free PMC article.
-
Heparan sulfate proteoglycans serve as alternative receptors for low affinity LCMV variants.PLoS Pathog. 2021 Oct 14;17(10):e1009996. doi: 10.1371/journal.ppat.1009996. eCollection 2021 Oct. PLoS Pathog. 2021. PMID: 34648606 Free PMC article.
-
Congenital viral infections of the brain: lessons learned from lymphocytic choriomeningitis virus in the neonatal rat.PLoS Pathog. 2007 Nov;3(11):e149. doi: 10.1371/journal.ppat.0030149. PLoS Pathog. 2007. PMID: 18052527 Free PMC article. Review.
-
Lymphocytic choriomeningitis virus injures the developing brain: effects and mechanisms.Pediatr Res. 2024 Jan;95(2):551-557. doi: 10.1038/s41390-023-02985-5. Epub 2024 Jan 5. Pediatr Res. 2024. PMID: 38182822 Review.
Cited by
-
A review of virus host factor discovery using CRISPR screening.mBio. 2024 Nov 13;15(11):e0320523. doi: 10.1128/mbio.03205-23. Epub 2024 Oct 18. mBio. 2024. PMID: 39422472 Free PMC article. Review.
-
U-73122, a phospholipase C inhibitor, impairs lymphocytic choriomeningitis virus virion infectivity.J Gen Virol. 2024 Dec;105(12):002060. doi: 10.1099/jgv.0.002060. J Gen Virol. 2024. PMID: 39688895 Free PMC article.
-
Systematic review and meta-analysis of genome-wide pooled CRISPR screens to identify host factors involved in influenza A virus infection.J Virol. 2024 May 14;98(5):e0185723. doi: 10.1128/jvi.01857-23. Epub 2024 Apr 3. J Virol. 2024. PMID: 38567969 Free PMC article.
-
Segmented, Negative-Sense RNA Viruses of Humans: Genetic Systems and Experimental Uses of Reporter Strains.Annu Rev Virol. 2023 Sep 29;10(1):261-282. doi: 10.1146/annurev-virology-111821-120445. Annu Rev Virol. 2023. PMID: 37774125 Free PMC article. Review.
-
CRISPR screens in Drosophila cells identify Vsg as a Tc toxin receptor.Nature. 2022 Oct;610(7931):349-355. doi: 10.1038/s41586-022-05250-7. Epub 2022 Sep 28. Nature. 2022. PMID: 36171290 Free PMC article.
References
-
- Radoshitzky SR, Buchmeier MJ, Charrel RN, Clegg JCS, Gonzalez J-PJ, Günther S, Hepojoki J, Kuhn JH, Lukashevich IS, Romanowski V, Salvato MS, Sironi M, Stenglein MD, de la Torre JC, ICTV Report Consortium . 2019. ICTV Virus Taxonomy Profile: arenaviridae. J Gen Virol 100:1200–1201. doi:10.1099/jgv.0.001280. - DOI - PubMed
-
- Stenglein MD, Sanchez-Migallon Guzman D, Garcia VE, Layton ML, Hoon-Hanks LL, Boback SM, Keel MK, Drazenovich T, Hawkins MG, DeRisi JL. 2017. Differential disease susceptibilities in experimentally reptarenavirus-infected boa constrictors and ball pythons. J Virol 91:e00451-17. doi:10.1128/JVI.00451-17. - DOI - PMC - PubMed
-
- Hepojoki J, Hepojoki S, Smura T, Szirovicza L, Dervas E, Prähauser B, Nufer L, Schraner EM, Vapalahti O, Kipar A, Hetzel U. 2018. Characterization of Haartman Institute snake virus-1 (HISV-1) and HISV-like viruses–the representatives of genus Hartmanivirus, family Arenaviridae. PLoS Pathog 14:e1007415. doi:10.1371/journal.ppat.1007415. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

