Profiling the molecular signature of satellite glial cells at the single cell level reveals high similarities between rodents and humans
- PMID: 35503034
- PMCID: PMC9522926
- DOI: 10.1097/j.pain.0000000000002628
Profiling the molecular signature of satellite glial cells at the single cell level reveals high similarities between rodents and humans
Abstract
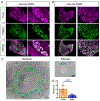
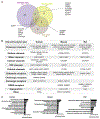
Peripheral sensory neurons located in dorsal root ganglia relay sensory information from the peripheral tissue to the brain. Satellite glial cells (SGCs) are unique glial cells that form an envelope completely surrounding each sensory neuron soma. This organization allows for close bidirectional communication between the neuron and its surrounding glial coat. Morphological and molecular changes in SGC have been observed in multiple pathological conditions such as inflammation, chemotherapy-induced neuropathy, viral infection, and nerve injuries. There is evidence that changes in SGC contribute to chronic pain by augmenting the neuronal activity in various rodent pain models. Satellite glial cells also play a critical role in axon regeneration. Whether findings made in rodent model systems are relevant to human physiology have not been investigated. Here, we present a detailed characterization of the transcriptional profile of SGC in mice, rats, and humans at the single cell level. Our findings suggest that key features of SGC in rodent models are conserved in humans. Our study provides the potential to leverage rodent SGC properties and identify potential targets in humans for the treatment of nerve injuries and alleviation of painful conditions.
Copyright © 2022 International Association for the Study of Pain.
Conflict of interest statement
CONFLICTS OF INTEREST
The authors declare no conflict of interest
Figures





References
-
- Barres BA, Smith SJ. Neurobiology. Cholesterol--making or breaking the synapse. Science 2001;294(5545):1296–1297. - PubMed
-
- Bogdanov P, Hernandez C, Corraliza L, Carvalho AR, Simo R. Effect of fenofibrate on retinal neurodegeneration in an experimental model of type 2 diabetes. Acta Diabetol 2015;52(1):113–122. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

