Function deficiency of GhOMT1 causes anthocyanidins over-accumulation and diversifies fibre colours in cotton (Gossypium hirsutum)
- PMID: 35503731
- PMCID: PMC9342615
- DOI: 10.1111/pbi.13832
Function deficiency of GhOMT1 causes anthocyanidins over-accumulation and diversifies fibre colours in cotton (Gossypium hirsutum)
Abstract
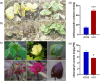
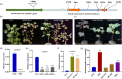
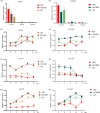
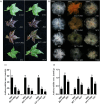
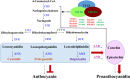
Naturally coloured cotton (NCC) fibres need little or no dyeing process in textile industry to low-carbon emission and are environment-friendly. Proanthocyanidins (PAs) and their derivatives were considered as the main components causing fibre coloration and made NCCs very popular and healthy, but the monotonous fibre colours greatly limit the wide application of NCCs. Here a G. hirsutum empurpled mutant (HS2) caused by T-DNA insertion is found to enhance the anthocyanidins biosynthesis and accumulate anthocyanidins in the whole plant. HPLC and LC/MS-ESI analysis confirmed the anthocyanidins methylation and peonidin, petunidin and malvidin formation are blocked. The deficiency of GhOMT1 in HS2 was associated with the activation of the anthocyanidin biosynthesis and the altered components of anthocyanidins. The transcripts of key genes in anthocyanidin biosynthesis pathway are significantly up-regulated in HS2, while transcripts of the genes for transport and decoration were at similar levels as in WT. To investigate the potential mechanism of GhOMT1 deficiency in cotton fibre coloration, HS2 mutant was crossed with NCCs. Surprisingly, offsprings of HS2 and NCCs enhanced PAs biosynthesis and increased PAs levels in their fibres from the accumulated anthocyanidins through up-regulated GhANR and GhLAR. As expected, multiple novel lines with improved fibre colours including orange red and navy blue were produced in their generations. Based on this work, a new strategy for breeding diversified NCCs was brought out by promoting PA biosynthesis. This work will help shed light on mechanisms of PA biosynthesis and bring out potential molecular breeding strategy to increase PA levels in NCCs.
Keywords: anthocyanidin methylation; fibre colour; flavonoid O-methyltransferase gene; naturally coloured cotton; proanthocyanidin biosynthesis.
© 2022 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
No conflict of interest declared.
Figures


References
-
- Akita, Y. , Kitamura, S. , Hase, Y. , Narumi, I. , Ishizaka, H. , Kondo, E. , Kameari, N. et al. (2011) Isolation and characterization of the fragrant cyclamen O‐methyltransferase involved in flower coloration. Planta, 234, 1127–1136. - PubMed
-
- Bassolino, L. , Zhang, Y. , Schoonbeek, H.J. , Kiferle, C. , Perata, P. and Martin, C. (2013) Accumulation of anthocyanins in tomato skin extends shelf life. New Phytol. 200, 650–655. - PubMed
-
- Berni, R. , Cai, G. , Hausman, J.‐F. and Guerriero, G. (2019) Plant fibers and phenolics: A review on their synthesis, analysis and combined use for biomaterials with new properties. Fibers, 7, 80.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

