A neutralizing antibody target in early HIV-1 infection was recapitulated in rhesus macaques immunized with the transmitted/founder envelope sequence
- PMID: 35503780
- PMCID: PMC9106183
- DOI: 10.1371/journal.ppat.1010488
A neutralizing antibody target in early HIV-1 infection was recapitulated in rhesus macaques immunized with the transmitted/founder envelope sequence
Abstract
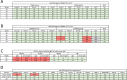
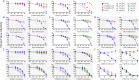
Transmitted/founder (T/F) HIV-1 envelope proteins (Envs) from infected individuals that developed neutralization breadth are likely to possess inherent features desirable for vaccine immunogen design. To explore this premise, we conducted an immunization study in rhesus macaques (RM) using T/F Env sequences from two human subjects, one of whom developed potent and broad neutralizing antibodies (Z1800M) while the other developed little to no neutralizing antibody responses (R66M) during HIV-1 infection. Using a DNA/MVA/protein immunization protocol, 10 RM were immunized with each T/F Env. Within each T/F Env group, the protein boosts were administered as either monomeric gp120 or stabilized trimeric gp140 protein. All vaccination regimens elicited high titers of antigen-specific IgG, and two animals that received monomeric Z1800M Env gp120 developed autologous neutralizing activity. Using early Env escape variants isolated from subject Z1800M as guides, the serum neutralizing activity of the two immunized RM was found to be dependent on the gp120 V5 region. Interestingly, the exact same residues of V5 were also targeted by a neutralizing monoclonal antibody (nmAb) isolated from the subject Z1800M early in infection. Glycan profiling and computational modeling of the Z1800M Env gp120 immunogen provided further evidence that the V5 loop is exposed in this T/F Env and was a dominant feature that drove neutralizing antibody targeting during infection and immunization. An expanded B cell clonotype was isolated from one of the neutralization-positive RM and nmAbs corresponding to this group demonstrated V5-dependent neutralization similar to both the RM serum and the human Z1800M nmAb. The results demonstrate that neutralizing antibody responses elicited by the Z1800M T/F Env in RM converged with those in the HIV-1 infected human subject, illustrating the potential of using immunogens based on this or other T/F Envs with well-defined immunogenicity as a starting point to drive breadth.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






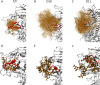






References
-
- Wagh K, Kreider EF, Li Y, Barbian HJ, Learn GH, Giorgi E, et al. Completeness of HIV-1 Envelope Glycan Shield at Transmission Determines Neutralization Breadth. Cell reports. 2018;25(4):893–908 e7. Epub 2018/10/26. doi: 10.1016/j.celrep.2018.09.087 ; PubMed Central PMCID: PMC6426304. - DOI - PMC - PubMed
-
- van den Kerkhof TL, Feenstra KA, Euler Z, van Gils MJ, Rijsdijk LW, Boeser-Nunnink BD, et al. HIV-1 envelope glycoprotein signatures that correlate with the development of cross-reactive neutralizing activity. Retrovirology. 2013;10(1):102. Epub 2013/09/26. doi: 10.1186/1742-4690-10-102 ; PubMed Central PMCID: PMC3849187. - DOI - PMC - PubMed
-
- Smith SA, Burton SL, Kilembe W, Lakhi S, Karita E, Price M, et al. Diversification in the HIV-1 Envelope Hyper-variable Domains V2, V4, and V5 and Higher Probability of Transmitted/Founder Envelope Glycosylation Favor the Development of Heterologous Neutralization Breadth. PLoS pathogens. 2016;12(11):e1005989. Epub 2016/11/17. doi: 10.1371/journal.ppat.1005989 ; PubMed Central PMCID: PMC5112890. - DOI - PMC - PubMed
-
- Townsley SM, Donofrio GC, Jian N, Leggat DJ, Dussupt V, Mendez-Rivera L, et al. B cell engagement with HIV-1 founder virus envelope predicts development of broadly neutralizing antibodies. Cell host & microbe. 2021;29(4):564–78 e9. Epub 2021/03/05. doi: 10.1016/j.chom.2021.01.016 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

