Alterations in homologous recombination repair genes in prostate cancer brain metastases
- PMID: 35504881
- PMCID: PMC9065149
- DOI: 10.1038/s41467-022-30003-5
Alterations in homologous recombination repair genes in prostate cancer brain metastases
Abstract
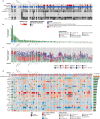
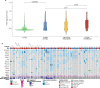
Improved survival rates for prostate cancer through more effective therapies have also led to an increase in the diagnosis of metastases to infrequent locations such as the brain. Here we investigate the repertoire of somatic genetic alterations present in brain metastases from 51 patients with prostate cancer brain metastases (PCBM). We highlight the clonal evolution occurring in PCBM and demonstrate an increased mutational burden, concomitant with an enrichment of the homologous recombination deficiency mutational signature in PCBM compared to non-brain metastases. Focusing on known pathogenic alterations within homologous recombination repair genes, we find 10 patients (19.6%) fulfilling the inclusion criteria used in the PROfound clinical trial, which assessed the efficacy of PARP inhibitors (PARPi) in homologous recombination deficient prostate cancer. Eight (15.7%) patients show biallelic loss of one of the 15 genes included in the trial, while 5 patients (9.8%) harbor pathogenic alterations in BRCA1/2 specifically. Uncovering these molecular features of PCBM may have therapeutic implications, suggesting the need of clinical trial enrollment of PCBM patients when evaluating potential benefit from PARPi.
© 2022. The Author(s).
Conflict of interest statement
S.A.T. and M.A.R. are co-authors (and included in the royalty streams) on the patent US7718369B2 issued to the University of Michigan and the Brigham and Women’s Hospital, on ETS gene fusions that have been licensed to Hologic/Gen-Probe Inc., who sublicensed rights to Roche/Ventana Medical Systems, and LynxDX. S.A.T. has served as a consultant for and received honoraria from Janssen, and Astellas/Medivation. S.A.T. has sponsored research agreements with Astellas/Medivation. S.A.T. is a cofounder of, prior consultant for, equity holder in, and employee of Strata Oncology. S.G. plays a consulting or advisory role to Astellas Pharma (Inst), Curevac (Inst), Novartis (Inst), Active Biotech (Inst), Bristol-Myers Squibb (Inst), Ferring (Inst), MaxiVax, Advanced Accelerator Applications, Roche, Janssen (Inst), Innocrin Pharma (Inst), Sanofi, Bayer (Inst), Orion Pharma GmbH, Clovis Oncology (Inst), Menarini Silicon Biosystems (Inst), MSD (Inst). S.G. is a co-author of the patent Method for biomarker (WO 3752009138392 A1). S.G. is an honorary member of Janssen and has also ties to Nektar, ProteoMediX. M.A.R. is on the Scientific Advisory Board of NeoGenomics, inc. C.K.Y.N. serves as a consultant for Repare Therapeutics USA. L.B. has served as an advisor for Janssen, Bayer, and Roche, has received honoraria from Janssen, and has sponsored research agreement with Novartis. The remaining authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

