Alternative splicing modulation by G-quadruplexes
- PMID: 35504902
- PMCID: PMC9065059
- DOI: 10.1038/s41467-022-30071-7
Alternative splicing modulation by G-quadruplexes
Abstract
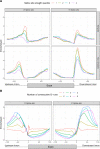
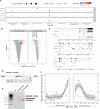
Alternative splicing is central to metazoan gene regulation, but the regulatory mechanisms are incompletely understood. Here, we show that G-quadruplex (G4) motifs are enriched ~3-fold near splice junctions. The importance of G4s in RNA is emphasised by a higher enrichment for the non-template strand. RNA-seq data from mouse and human neurons reveals an enrichment of G4s at exons that were skipped following depolarisation induced by potassium chloride. We validate the formation of stable RNA G4s for three candidate splice sites by circular dichroism spectroscopy, UV-melting and fluorescence measurements. Moreover, we find that sQTLs are enriched at G4s, and a minigene experiment provides further support for their role in promoting exon inclusion. Analysis of >1,800 high-throughput experiments reveals multiple RNA binding proteins associated with G4s. Finally, exploration of G4 motifs across eleven species shows strong enrichment at splice sites in mammals and birds, suggesting an evolutionary conserved splice regulatory mechanism.
© 2022. The Author(s).
Conflict of interest statement
I.G.S. and M.H. are founders of Neomer Diagnostics. E.A.M. is a founder and director of STORM Therapeutics. The remaining authors declare no competing interests.
Figures





References
-
- Oesterreich, F. C., Bowne-Anderson, H. & Howard, J. The contribution of alternative splicing probability to the coding expansion of the genome. bioRxiv10.1101/048124 (2016).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

