DNA methylation signature of chronic low-grade inflammation and its role in cardio-respiratory diseases
- PMID: 35504910
- PMCID: PMC9065016
- DOI: 10.1038/s41467-022-29792-6
DNA methylation signature of chronic low-grade inflammation and its role in cardio-respiratory diseases
Abstract
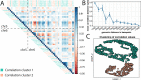
We performed a multi-ethnic Epigenome Wide Association study on 22,774 individuals to describe the DNA methylation signature of chronic low-grade inflammation as measured by C-Reactive protein (CRP). We find 1,511 independent differentially methylated loci associated with CRP. These CpG sites show correlation structures across chromosomes, and are primarily situated in euchromatin, depleted in CpG islands. These genomic loci are predominantly situated in transcription factor binding sites and genomic enhancer regions. Mendelian randomization analysis suggests altered CpG methylation is a consequence of increased blood CRP levels. Mediation analysis reveals obesity and smoking as important underlying driving factors for changed CpG methylation. Finally, we find that an activated CpG signature significantly increases the risk for cardiometabolic diseases and COPD.
© 2022. The Author(s).
Conflict of interest statement
H.J.G. has received travel grants and speakers honoraria from Fresenius Medical Care, Neuraxpharm, Servier, and Janssen Cilag as well as research funding from Fresenius Medical Care. All other study authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- 75N92021D00006/HL/NHLBI NIH HHS/United States
- N01HC85082/HL/NHLBI NIH HHS/United States
- MR/K026992/1/MRC_/Medical Research Council/United Kingdom
- HHSN268201700005I/HL/NHLBI NIH HHS/United States
- HHSN268200800007C/HL/NHLBI NIH HHS/United States
- MC_UU_00011/5/MRC_/Medical Research Council/United Kingdom
- G1001357/MRC_/Medical Research Council/United Kingdom
- N01HC85080/HL/NHLBI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- BB/S020845/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 HL086694/HL/NHLBI NIH HHS/United States
- 16/136/68/DH_/Department of Health/United Kingdom
- R01 HL131136/HL/NHLBI NIH HHS/United States
- MR/R023484/1/MRC_/Medical Research Council/United Kingdom
- R01 AG023629/AG/NIA NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- R01 NS087541/NS/NINDS NIH HHS/United States
- WT081878MA/WT_/Wellcome Trust/United Kingdom
- R01 HL103612/HL/NHLBI NIH HHS/United States
- K08 HL116640/HL/NHLBI NIH HHS/United States
- HHSN268201700001I/HL/NHLBI NIH HHS/United States
- WT202786/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- R01 HL092111/HL/NHLBI NIH HHS/United States
- 217065/Z/19/Z/WT_/Wellcome Trust/United Kingdom
- MR/S03532X/1/MRC_/Medical Research Council/United Kingdom
- HHSN268201700004I/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- 212945/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- N01HC55222/HL/NHLBI NIH HHS/United States
- R01 HL111089/HL/NHLBI NIH HHS/United States
- BBI025751/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- N01HC85079/HL/NHLBI NIH HHS/United States
- R01 HL059367/HL/NHLBI NIH HHS/United States
- MR/M01311/1/MRC_/Medical Research Council/United Kingdom
- N01HC85083/HL/NHLBI NIH HHS/United States
- P30 ES030285/ES/NIEHS NIH HHS/United States
- HHSN268201700002I/HL/NHLBI NIH HHS/United States
- U01 HG004402/HG/NHGRI NIH HHS/United States
- N01HC85086/HL/NHLBI NIH HHS/United States
- MR/S019669/1/MRC_/Medical Research Council/United Kingdom
- HHSN268201700003I/HL/NHLBI NIH HHS/United States
- WT092830/Z/10/Z/WT_/Wellcome Trust/United Kingdom
- R01 HL087641/HL/NHLBI NIH HHS/United States
- RE/18/4/34215/BHF_/British Heart Foundation/United Kingdom
- U01 HL130114/HL/NHLBI NIH HHS/United States
- RG/14/5/30893/BHF_/British Heart Foundation/United Kingdom
- R01 HL087652/HL/NHLBI NIH HHS/United States
- G0601653/MRC_/Medical Research Council/United Kingdom
- R01 HL116747/HL/NHLBI NIH HHS/United States
- MR/L01632X/1/MRC_/Medical Research Council/United Kingdom
- N01HC85081/HL/NHLBI NIH HHS/United States
- BB/T019980/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- UL1 TR001881/TR/NCATS NIH HHS/United States
- BB/I025263/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_UU_00006/2/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

