Longitudinal multi-omics analyses link gut microbiome dysbiosis with recurrent urinary tract infections in women
- PMID: 35505248
- PMCID: PMC9136705
- DOI: 10.1038/s41564-022-01107-x
Longitudinal multi-omics analyses link gut microbiome dysbiosis with recurrent urinary tract infections in women
Abstract
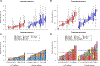
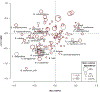
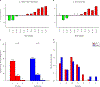
Recurrent urinary tract infections (rUTIs) are a major health burden worldwide, with history of infection being a significant risk factor. While the gut is a known reservoir for uropathogenic bacteria, the role of the microbiota in rUTI remains unclear. We conducted a year-long study of women with (n = 15) and without (n = 16) history of rUTI, from whom we collected urine, blood and monthly faecal samples for metagenomic and transcriptomic interrogation. During the study 24 UTIs were reported, with additional samples collected during and after infection. The gut microbiome of individuals with a history of rUTI was significantly depleted in microbial richness and butyrate-producing bacteria compared with controls, reminiscent of other inflammatory conditions. However, Escherichia coli gut and bladder populations were comparable between cohorts in both relative abundance and phylogroup. Transcriptional analysis of peripheral blood mononuclear cells revealed expression profiles indicative of differential systemic immunity between cohorts. Altogether, these results suggest that rUTI susceptibility is in part mediated through the gut-bladder axis, comprising gut dysbiosis and differential immune response to bacterial bladder colonization, manifesting in symptoms.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures

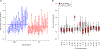


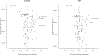


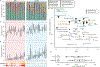


Comment in
-
Gut-bladder axis in recurrent UTI.Nat Microbiol. 2022 May;7(5):601-602. doi: 10.1038/s41564-022-01113-z. Nat Microbiol. 2022. PMID: 35505247 No abstract available.
-
Infection and Inflammation of the Genitourinary Tract.J Urol. 2022 Nov;208(5):1141-1142. doi: 10.1097/JU.0000000000002906. Epub 2022 Aug 11. J Urol. 2022. PMID: 35950377 No abstract available.
References
-
- Hooton TM, et al. A prospective study of risk factors for symptomatic urinary tract infection in young women. N Engl J Med 335, 468–474 (1996). - PubMed
-
- Yamamoto S, et al. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J Urol 157, 1127–1129 (1997). - PubMed
-
- Nielsen KL, Dynesen P, Larsen P & Frimodt-Moller N Faecal Escherichia coli from patients with E. coli urinary tract infection and healthy controls who have never had a urinary tract infection. J Med Microbiol 63, 582–589 (2014). - PubMed
-
- Jantunen ME, Saxen H, Lukinmaa S, Ala-Houhala M & Siitonen A Genomic identity of pyelonephritogenic Escherichia coli isolated from blood, urine and faeces of children with urosepsis. J Med Microbiol 50, 650–652 (2001). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

