Increased accuracy of genomic predictions for growth under chronic thermal stress in rainbow trout by prioritizing variants from GWAS using imputed sequence data
- PMID: 35505881
- PMCID: PMC9046923
- DOI: 10.1111/eva.13240
Increased accuracy of genomic predictions for growth under chronic thermal stress in rainbow trout by prioritizing variants from GWAS using imputed sequence data
Abstract
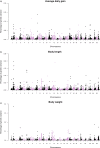
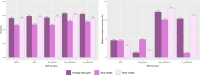
Through imputation of genotypes, genome-wide association study (GWAS) and genomic prediction (GP) using whole-genome sequencing (WGS) data are cost-efficient and feasible in aquaculture breeding schemes. The objective was to dissect the genetic architecture of growth traits under chronic heat stress in rainbow trout (Oncorhynchus mykiss) and to assess the accuracy of GP based on imputed WGS and different preselected single nucleotide polymorphism (SNP) arrays. A total of 192 and 764 fish challenged to a heat stress experiment for 62 days were genotyped using a customized 1 K and 26 K SNP panels, respectively, and then, genotype imputation was performed from a low-density chip to WGS using 102 parents (36 males and 66 females) as the reference population. Imputed WGS data were used to perform GWAS and test GP accuracy under different preselected SNP scenarios. Heritability was estimated for body weight (BW), body length (BL) and average daily gain (ADG). Estimates using imputed WGS data ranged from 0.33 ± 0.05 to 0.55 ± 0.05 for growth traits under chronic heat stress. GWAS revealed that the top five cumulatively SNPs explained a maximum of 0.94%, 0.86% and 0.51% of genetic variance for BW, BL and ADG, respectively. Some important functional candidate genes associated with growth-related traits were found among the most important SNPs, including signal transducer and activator of transcription 5B and 3 (STAT5B and STAT3, respectively) and cytokine-inducible SH2-containing protein (CISH). WGS data resulted in a slight increase in prediction accuracy compared with pedigree-based method, whereas preselected SNPs based on the top GWAS hits improved prediction accuracies, with values ranging from 1.2 to 13.3%. Our results support the evidence of the polygenic nature of growth traits when measured under heat stress. The accuracies of GP can be improved using preselected variants from GWAS, and the use of WGS marginally increases prediction accuracy.
Keywords: GWAS; accuracy; genomic predictions; heat stress; rainbow trout; whole‐genome sequence.
© 2021 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
Both authors declare no conflict of interest.
Figures
Similar articles
-
Strategies for Obtaining and Pruning Imputed Whole-Genome Sequence Data for Genomic Prediction.Front Genet. 2019 Jul 17;10:673. doi: 10.3389/fgene.2019.00673. eCollection 2019. Front Genet. 2019. PMID: 31379929 Free PMC article.
-
Genomic prediction based on preselected single-nucleotide polymorphisms from genome-wide association study and imputed whole-genome sequence data annotation for growth traits in Duroc pigs.Evol Appl. 2024 Feb 15;17(2):e13651. doi: 10.1111/eva.13651. eCollection 2024 Feb. Evol Appl. 2024. PMID: 38362509 Free PMC article.
-
GWAS on Imputed Whole-Genome Sequence Variants Reveal Genes Associated with Resistance to Piscirickettsia salmonis in Rainbow Trout (Oncorhynchus mykiss).Genes (Basel). 2022 Dec 30;14(1):114. doi: 10.3390/genes14010114. Genes (Basel). 2022. PMID: 36672855 Free PMC article.
-
Genome-wide association study and genomic prediction for intramuscular fat content in Suhuai pigs using imputed whole-genome sequencing data.Evol Appl. 2022 Oct 24;15(12):2054-2066. doi: 10.1111/eva.13496. eCollection 2022 Dec. Evol Appl. 2022. PMID: 36540634 Free PMC article.
-
Evaluation of measures of correctness of genotype imputation in the context of genomic prediction: a review of livestock applications.Animal. 2014 Nov;8(11):1743-53. doi: 10.1017/S1751731114001803. Epub 2014 Jul 21. Animal. 2014. PMID: 25045914 Review.
Cited by
-
Effects of common full-sib families on accuracy of genomic prediction for tagging weight in striped catfish Pangasianodon hypophthalmus.Front Genet. 2023 Jan 4;13:1081246. doi: 10.3389/fgene.2022.1081246. eCollection 2022. Front Genet. 2023. PMID: 36685869 Free PMC article.
-
Genetic parameters and genome-wide association for milk production traits and somatic cell score in different lactation stages of Shanghai Holstein population.Front Genet. 2022 Sep 5;13:940650. doi: 10.3389/fgene.2022.940650. eCollection 2022. Front Genet. 2022. PMID: 36134029 Free PMC article.
-
Strategies to improve the accuracy and reduce costs of genomic prediction in aquaculture species.Evol Appl. 2021 Jul 17;15(4):578-590. doi: 10.1111/eva.13262. eCollection 2022 Apr. Evol Appl. 2021. PMID: 35505889 Free PMC article.
-
Enhancing genomic prediction in Arabidopsis thaliana with optimized SNP subset by leveraging gene ontology priors and bin-based combinatorial optimization.Front Bioinform. 2025 Jun 18;5:1607119. doi: 10.3389/fbinf.2025.1607119. eCollection 2025. Front Bioinform. 2025. PMID: 40607015 Free PMC article.
-
Genome-Wide Association Study Reveals SNPs and Candidate Genes Related to Growth and Body Shape in Bighead Carp (Hypophthalmichthys nobilis).Mar Biotechnol (NY). 2022 Dec;24(6):1138-1147. doi: 10.1007/s10126-022-10176-2. Epub 2022 Nov 9. Mar Biotechnol (NY). 2022. PMID: 36350467
References
-
- Abdollahi‐Arpanahi, R. , Nejati‐Javaremi, A. , Pakdel, A. , Moradi‐Shahrbabak, M. , Morota, G. , Valente, B. D. , Kranis, A. , Rosa, G. J. M. , & Gianola, D. (2014). Effect of allele frequencies, effect sizes and number of markers on prediction of quantitative traits in chickens. Journal of Animal Breeding and Genetics, 131(2), 123–133. 10.1111/jbg.12075 - DOI - PubMed
-
- Aguilar, I. , Misztal, I. , Johnson, D. L. , Legarra, A. , Tsuruta, S. , & Lawlor, T. J. (2010). Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. Journal of Dairy Science, 93(2), 743–752. 10.3168/jds.2009-2730 - DOI - PubMed
-
- Barría, A. , Christensen, K. A. , Yoshida, G. M. , Correa, K. , Jedlicki, A. , Lhorente, J. P. , Davidson, W. S. , & Yáñez, J. M. (2018). Genomic predictions and genome‐wide association study of resistance against Piscirickettsia salmonis in Coho salmon (Oncorhynchus kisutch) using ddRAD sequencing. G3: Genes, Genomes, Genetics, 8, 1183–1194. - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

