The Impact of Activity-Based Protein Profiling in Malaria Drug Discovery
- PMID: 35506504
- PMCID: PMC9401580
- DOI: 10.1002/cmdc.202200174
The Impact of Activity-Based Protein Profiling in Malaria Drug Discovery
Abstract
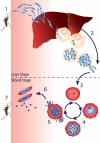
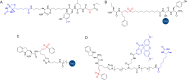
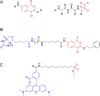
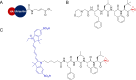
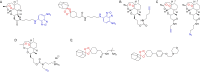
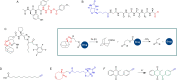
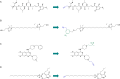
Activity-based protein profiling (ABPP) is an approach used at the interface of chemical biology and proteomics that uses small molecular probes to provide dynamic fingerprints of enzymatic activity in complex proteomes. Malaria is a disease caused by Plasmodium parasites with a significant death burden and for which new therapies are actively being sought. Here, we compile the main achievements from ABPP studies in malaria and highlight the probes used and the different downstream platforms for data analysis. ABPP has excelled at studying Plasmodium cysteine proteases and serine hydrolase families, the targeting of the proteasome and metabolic pathways, and in the deconvolution of targets and mechanisms of known antimalarials. Despite the major impact in the field, many antimalarials and enzymatic families in Plasmodium remain to be studied, which suggests ABPP will be an evergreen technique in the field.
Keywords: Drug Discovery; Fluorescent probes; Malaria; Mass spectrometry; Proteomics.
© 2022 The Authors. ChemMedChem published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Kidd D., Liu Y., Cravatt B. F., Biochemistry 2001, 40, 4005–4015. - PubMed
-
- Speers A. E., Cravatt B. F., J. Am. Chem. Soc. 2005, 127, 10018–10019; N. Jessani, S. Niessen, B. Q. Wei, M. Nicolau, M. Humphrey, Y. Ji, W. Han, D. Y. Noh, J. R. Yates, 3rd, S. S. Jeffrey, B. F.
- Cravatt, Nat. Methods 2005, 2, 691–697. - PubMed
-
- Sadaghiani A. M., Verhelst S. H., Bogyo M., Curr. Opin. Chem. Biol. 2007, 11, 20–28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

