ICTV Virus Taxonomy Profile: Potyviridae 2022
- PMID: 35506996
- PMCID: PMC12642016
- DOI: 10.1099/jgv.0.001738
ICTV Virus Taxonomy Profile: Potyviridae 2022
Abstract
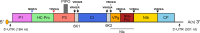
The family Potyviridae includes plant viruses with single-stranded, positive-sense RNA genomes of 8-11 kb and flexuous filamentous particles 650-950 nm long and 11-20 nm wide. Genera in the family are distinguished by the host range, genomic features and phylogeny of the member viruses. Most genomes are monopartite, but those of members of the genus Bymovirus are bipartite. Some members cause serious disease epidemics in cultivated plants. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Potyviridae, which is available at ictv.global/report/potyviridae.
Keywords: ICTV Profile; Potyviridae; taxonomy.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

