Whole-genome resequencing reveals domestication and signatures of selection in Ujimqin, Sunit, and Wu Ranke Mongolian sheep breeds
- PMID: 35507861
- PMCID: PMC9449395
- DOI: 10.5713/ab.21.0569
Whole-genome resequencing reveals domestication and signatures of selection in Ujimqin, Sunit, and Wu Ranke Mongolian sheep breeds
Abstract
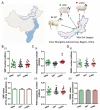
Objective: The current study aimed to perform whole-genome resequencing of Chinese indigenous Mongolian sheep breeds including Ujimqin, Sunit, and Wu Ranke sheep breeds (UJMQ, SNT, WRK) and deeply analyze genetic variation, population structure, domestication, and selection for domestication traits among these Mongolian sheep breeds.
Methods: Blood samples were collected from a total of 60 individuals comprising 20 WRK, 20 UJMQ, and 20 SNT. For genome sequencing, about 1.5 μg of genomic DNA was used for library construction with an insert size of about 350 bp. Pair-end sequencing were performed on Illumina NovaSeq platform, with the read length of 150 bp at each end. We then investigated the domestication and signatures of selection in these sheep breeds.
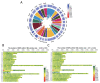
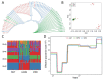
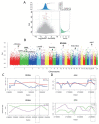
Results: According to the population and demographic analyses, WRK and SNT populations were very similar, which were different from UJMQ populations. Genome wide association study identified 468 and 779 significant loci from SNT vs UJMQ, and UJMQ vs WRK, respectively. However, only 3 loci were identified from SNT vs WRK. Genomic comparison and selective sweep analysis among these sheep breeds suggested that genes associated with regulation of secretion, metabolic pathways including estrogen metabolism and amino acid metabolism, and neuron development have undergone strong selection during domestication.
Conclusion: Our findings will facilitate the understanding of Chinese indigenous Mongolian sheep breeds domestication and selection for complex traits and provide a valuable genomic resource for future studies of sheep and other domestic animal breeding.
Keywords: Domestication; Metabolism; Selection; Sheep; Whole-genome Resequencing.
Conflict of interest statement
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

