Curcumin inhibits spike protein of new SARS-CoV-2 variant of concern (VOC) Omicron, an in silico study
- PMID: 35508082
- PMCID: PMC9044632
- DOI: 10.1016/j.compbiomed.2022.105552
Curcumin inhibits spike protein of new SARS-CoV-2 variant of concern (VOC) Omicron, an in silico study
Abstract
Background: Omicron (B.1.1.529), a variant of SARS-CoV-2 is currently spreading globally as a dominant strain. Due to multiple mutations at its Spike protein, including 15 amino acid substitutions at the receptor binding domain (RBD), Omicron is a variant of concern (VOC) and capable of escaping vaccine generated immunity. So far, no specific treatment regime is suggested for this VOC.
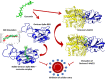
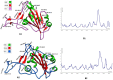
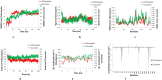
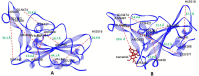
Methods: The three-dimensional structure of the Spike RBD domain of Omicron variant was constructed by incorporating 15 amino acid substitutions to the Native Spike (S) structure and structural changes were compared that of the Native S. Seven phytochemicals namely Allicin, Capsaicin, Cinnamaldehyde, Curcumin, Gingerol, Piperine, and Zingeberene were docked with Omicron S protein and Omicron S-hACE2 complex. Further, molecular dynamic simulation was performed between Crcumin and Omicron S protein to evaluate the structural stability of the complex in the physiological environment and compared with that of the control drug Chloroquine.
Results: Curcumin, among seven phytochemicals, was found to have the most substantial inhibitory potential with Omicron S protein. Further, it was found that curcumin could disrupt the Omicron S-hACE2 complex. The molecular dynamic simulation demonstrated that Curcumin could form a stable structure with Omicron S in the physiological environment.
Conclusion: To conclude, Curcumin can be considered as a potential therapeutic agent against the highly infectious Omicron variant of SARS-CoV-2.
Keywords: COVID-19; Coronavirus; In silico study; Omicron; Spike-RBD.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Authors declare that there is no conflict of interest.
Figures

Similar articles
-
In silico study of some selective phytochemicals against a hypothetical SARS-CoV-2 spike RBD using molecular docking tools.Comput Biol Med. 2021 Oct;137:104818. doi: 10.1016/j.compbiomed.2021.104818. Epub 2021 Aug 28. Comput Biol Med. 2021. PMID: 34481181 Free PMC article.
-
In silico structural inhibition of ACE-2 binding site of SARS-CoV-2 and SARS-CoV-2 omicron spike protein by lectin antiviral dyad system to treat COVID-19.Drug Dev Ind Pharm. 2022 Oct;48(10):539-551. doi: 10.1080/03639045.2022.2137196. Epub 2022 Oct 27. Drug Dev Ind Pharm. 2022. PMID: 36250723
-
In Silico Genome Analysis Reveals the Evolution and Potential Impact of SARS-CoV-2 Omicron Structural Changes on Host Immune Evasion and Antiviral Therapeutics.Viruses. 2022 Nov 6;14(11):2461. doi: 10.3390/v14112461. Viruses. 2022. PMID: 36366559 Free PMC article.
-
Omicron (B.1.1.529) - variant of concern - molecular profile and epidemiology: a mini review.Eur Rev Med Pharmacol Sci. 2021 Dec;25(24):8019-8022. doi: 10.26355/eurrev_202112_27653. Eur Rev Med Pharmacol Sci. 2021. PMID: 34982466 Review.
-
Literature Review of Omicron: A Grim Reality Amidst COVID-19.Microorganisms. 2022 Feb 16;10(2):451. doi: 10.3390/microorganisms10020451. Microorganisms. 2022. PMID: 35208905 Free PMC article. Review.
Cited by
-
Natural products as a source of Coronavirus entry inhibitors.Front Cell Infect Microbiol. 2024 Feb 21;14:1353971. doi: 10.3389/fcimb.2024.1353971. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38449827 Free PMC article. Review.
-
Curcumin as an antiviral agent and immune-inflammatory modulator in COVID-19: A scientometric analysis.Heliyon. 2023 Nov 2;9(11):e21648. doi: 10.1016/j.heliyon.2023.e21648. eCollection 2023 Nov. Heliyon. 2023. PMID: 38027776 Free PMC article.
-
Evaluation of therapeutic potentials of selected phytochemicals against Nipah virus, a multi-dimensional in silico study.3 Biotech. 2023 Jun;13(6):174. doi: 10.1007/s13205-023-03595-y. Epub 2023 May 10. 3 Biotech. 2023. PMID: 37180429 Free PMC article.
-
Clinical Approach to Post-acute Sequelae After COVID-19 Infection and Vaccination.Cureus. 2023 Nov 21;15(11):e49204. doi: 10.7759/cureus.49204. eCollection 2023 Nov. Cureus. 2023. PMID: 38024037 Free PMC article. Review.
-
Therapeutic potential of curcumin in ARDS and COVID-19.Clin Exp Pharmacol Physiol. 2023 Apr;50(4):267-276. doi: 10.1111/1440-1681.13744. Epub 2023 Jan 10. Clin Exp Pharmacol Physiol. 2023. PMID: 36480131 Free PMC article. Review.
References
-
- WHO Coronavirus (COVID-19) Dashboard n.d. https://covid19.who.int (accessed January 27, 2022).
-
- INSACOG | Department of Biotechnology n.d. https://dbtindia.gov.in/insacog (accessed January 27, 2022).
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

