Walking a tightrope: The complex balancing act of R-loops in genome stability
- PMID: 35508167
- PMCID: PMC9233011
- DOI: 10.1016/j.molcel.2022.04.014
Walking a tightrope: The complex balancing act of R-loops in genome stability
Abstract
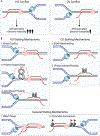
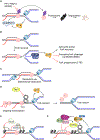
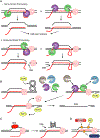
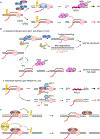
Although transcription is an essential cellular process, it is paradoxically also a well-recognized cause of genomic instability. R-loops, non-B DNA structures formed when nascent RNA hybridizes to DNA to displace the non-template strand as single-stranded DNA (ssDNA), are partially responsible for this instability. Yet, recent work has begun to elucidate regulatory roles for R-loops in maintaining the genome. In this review, we discuss the cellular contexts in which R-loops contribute to genomic instability, particularly during DNA replication and double-strand break (DSB) repair. We also summarize the evidence that R-loops participate as an intermediate during repair and may influence pathway choice to preserve genomic integrity. Finally, we discuss the immunogenic potential of R-loops and highlight their links to disease should they become pathogenic.
Keywords: DNA damage; R-loop pathology; R-loops; RNA-DNA hybrid; double-strand break repair; genome stability; replication stress.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Karlene A. Cimprich is a member of the Advisory Board for Molecular Cell.
Figures



References
-
- Aguilera A, and Gómez-González B (2008). Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet 9, 204–217. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

