Lipid exposure activates gene expression changes associated with estrogen receptor negative breast cancer
- PMID: 35508495
- PMCID: PMC9068822
- DOI: 10.1038/s41523-022-00422-0
Lipid exposure activates gene expression changes associated with estrogen receptor negative breast cancer
Erratum in
-
Author Correction: Lipid exposure activates gene expression changes associated with estrogen receptor negative breast cancer.NPJ Breast Cancer. 2023 Mar 13;9(1):11. doi: 10.1038/s41523-023-00516-3. NPJ Breast Cancer. 2023. PMID: 36914690 Free PMC article. No abstract available.
Abstract
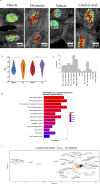
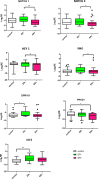
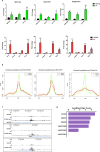
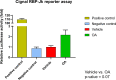
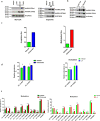
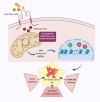
Improved understanding of local breast biology that favors the development of estrogen receptor negative (ER-) breast cancer (BC) would foster better prevention strategies. We have previously shown that overexpression of specific lipid metabolism genes is associated with the development of ER- BC. We now report results of exposure of MCF-10A and MCF-12A cells, and mammary organoids to representative medium- and long-chain polyunsaturated fatty acids. This exposure caused a dynamic and profound change in gene expression, accompanied by changes in chromatin packing density, chromatin accessibility, and histone posttranslational modifications (PTMs). We identified 38 metabolic reactions that showed significantly increased activity, including reactions related to one-carbon metabolism. Among these reactions are those that produce S-adenosyl-L-methionine for histone PTMs. Utilizing both an in-vitro model and samples from women at high risk for ER- BC, we show that lipid exposure engenders gene expression, signaling pathway activation, and histone marks associated with the development of ER- BC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

