North Asian population relationships in a global context
- PMID: 35508562
- PMCID: PMC9068624
- DOI: 10.1038/s41598-022-10706-x
North Asian population relationships in a global context
Abstract
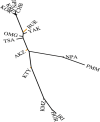
Population genetic studies of North Asian ethnic groups have focused on genetic variation of sex chromosomes and mitochondria. Studies of the extensive variation available from autosomal variation have appeared infrequently. We focus on relationships among population samples using new North Asia microhaplotype data. We combined genotypes from our laboratory on 58 microhaplotypes, distributed across 18 autosomes, on 3945 individuals from 75 populations with corresponding data extracted for 26 populations from the Thousand Genomes consortium and for 22 populations from the GenomeAsia 100 K project. A total of 7107 individuals in 122 total populations are analyzed using STRUCTURE, Principal Component Analysis, and phylogenetic tree analyses. North Asia populations sampled in Mongolia include: Buryats, Mongolians, Altai Kazakhs, and Tsaatans. Available Siberians include samples of Yakut, Khanty, and Komi Zyriane. Analyses of all 122 populations confirm many known relationships and show that most populations from North Asia form a cluster distinct from all other groups. Refinement of analyses on smaller subsets of populations reinforces the distinctiveness of North Asia and shows that the North Asia cluster identifies a region that is ancestral to Native Americans.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

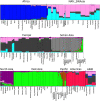

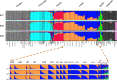

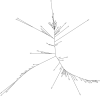
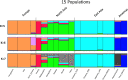

References
-
- Kidd KK, et al. Microhaplotype loci are a powerful new type of forensic marker. Forensic Sci. Int. Genet. Suppl. Ser. 2013;4:e123–e124. doi: 10.1016/j.fsigss.2013.10.063. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

