The mechanism of activation of MEK1 by B-Raf and KSR1
- PMID: 35508574
- PMCID: PMC9068654
- DOI: 10.1007/s00018-022-04296-0
The mechanism of activation of MEK1 by B-Raf and KSR1
Abstract
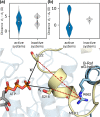
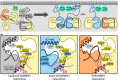
MEK1 interactions with B-Raf and KSR1 are key steps in Ras/Raf/MEK/ERK signaling. Despite this, vital mechanistic details of how these execute signal transduction are still enigmatic. Among these is why, despite B-Raf and KSR1 kinase domains similarity, the B-Raf/MEK1 and KSR1/MEK1 complexes have distinct contributions to MEK1 activation, and broadly, what is KSR1's role. Our molecular dynamics simulations clarify these still unresolved ambiguities. Our results reveal that the proline-rich (P-rich) loop of MEK1 plays a decisive role in MEK1 activation loop (A-loop) phosphorylation. In the inactive B-Raf/MEK1 heterodimer, the collapsed A-loop of B-Raf interacts with the P-rich loop and A-loop of MEK1, minimizing MEK1 A-loop fluctuation and preventing it from phosphorylation. In the active B-Raf/MEK1 heterodimer, the P-rich loop moves in concert with the A-loop of B-Raf as it extends. This reduces the number of residues interacting with MEK1 A-loop, allowing increased A-loop fluctuation, and bringing Ser222 closer to ATP for phosphorylation. B-Raf αG-helix Arg662 promotes MEK1 activation by orienting Ser218 towards ATP. In KSR1/MEK1, the KSR1 αG-helix has Ala826 in place of B-Raf Arg662. This difference results in much fewer interactions between KSR1 αG-helix and MEK1 A-loop, thus a more flexible A-loop. We postulate that if KSR1 were to adopt an active configuration with an extended A-loop as seen in other protein kinases, then the MEK1 P-rich loop would extend in a similar manner, as seen in the active B-Raf/MEK1 heterodimer. This would result in highly flexible MEK1 A-loop, and KSR1 functioning as an active, B-Raf-like, kinase.
Keywords: Assemblies; Autoinhibition; Cancer; ERK; KSR; MAPK; MD simulations.
© 2022. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

