Intermittent PI3Kδ inhibition sustains anti-tumour immunity and curbs irAEs
- PMID: 35508656
- PMCID: PMC9132770
- DOI: 10.1038/s41586-022-04685-2
Intermittent PI3Kδ inhibition sustains anti-tumour immunity and curbs irAEs
Abstract
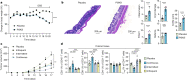
Phosphoinositide 3-kinase δ (PI3Kδ) has a key role in lymphocytes, and inhibitors that target this PI3K have been approved for treatment of B cell malignancies1-3. Although studies in mouse models of solid tumours have demonstrated that PI3Kδ inhibitors (PI3Kδi) can induce anti-tumour immunity4,5, its effect on solid tumours in humans remains unclear. Here we assessed the effects of the PI3Kδi AMG319 in human patients with head and neck cancer in a neoadjuvant, double-blind, placebo-controlled randomized phase II trial (EudraCT no. 2014-004388-20). PI3Kδ inhibition decreased the number of tumour-infiltrating regulatory T (Treg) cells and enhanced the cytotoxic potential of tumour-infiltrating T cells. At the tested doses of AMG319, immune-related adverse events (irAEs) required treatment to be discontinued in 12 out of 21 of patients treated with AMG319, suggestive of systemic effects on Treg cells. Accordingly, in mouse models, PI3Kδi decreased the number of Treg cells systemically and caused colitis. Single-cell RNA-sequencing analysis revealed a PI3Kδi-driven loss of tissue-resident colonic ST2 Treg cells, accompanied by expansion of pathogenic T helper 17 (TH17) and type 17 CD8+ T (TC17) cells, which probably contributed to toxicity; this points towards a specific mode of action for the emergence of irAEs. A modified treatment regimen with intermittent dosing of PI3Kδi in mouse models led to a significant decrease in tumour growth without inducing pathogenic T cells in colonic tissue, indicating that alternative dosing regimens might limit toxicity.
© 2022. The Author(s).
Conflict of interest statement
G.F. is an employee of Amgen Inc. B.V. is a consultant for iOnctura (Geneva, Switzerland), Venthera (Palo Alto, US) and Olema Pharmaceuticals (San Francisco, US) and has received speaker fees from Gilead (Foster City, US). K.O. has received consultancy fees from iOnctura, Macomics, Gilead Sciences and Karus Therapeutics and has received research funding from GSK. M.K. is on the scientific advisory board of Prometheus. C.H.O. led the clinical trial of AMG319 with funding by Cancer Research UK, Amgen provided clinical grade compound free of charge for this trial. All other authors declare no conflicts of interest.
Figures

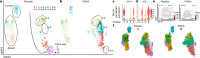







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

