Gut microbiota-derived short chain fatty acids are potential mediators in gut inflammation
- PMID: 35510031
- PMCID: PMC9040132
- DOI: 10.1016/j.aninu.2021.11.005
Gut microbiota-derived short chain fatty acids are potential mediators in gut inflammation
Abstract
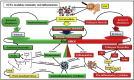
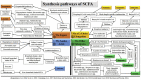
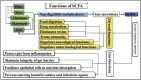
Gut inflammation is a challenging concern in humans and animals, which disturbs normal growth and leads to severe bowel diseases. Short chain fatty acids (SCFA) are the gut microbiota metabolites produced from fermentation of non-digestible carbohydrates, and have been reported to modulate gut inflammation. SCFA have been implicated as the potential therapeutic bioactive molecules for gut inflammatory diseases, and could be an alternative to antibiotic growth promoters (AGP). In this review, the existing knowledge about the types of SCFA, the related gut microbes producing SCFA, the roles of SCFA in maintaining gut homeostasis, and how SCFA modulate gut inflammation is summarized. The therapeutic application of SCFA in the treatment of inflammatory bowel disease (IBD) is also highlighted.
Keywords: Gut homeostasis; Gut inflammation; Gut microbiota; Inflammatory bowel disease; Short chain fatty acid.
© 2022 Chinese Association of Animal Science and Veterinary Medicine. Publishing services by Elsevier B.V. on behalf of KeAi Communications Co. Ltd.
Conflict of interest statement
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Figures




References
-
- Akhtar M., Shaukat A., Zahoor A., Chen Y., Wang Y., Yang M., et al. Hederacoside-c inhibition of staphylococcus aureus-induced mastitis via tlr2 & tlr4 and their downstream signaling nf-κb and mapks pathways in vivo and in vitro. Inflammation. 2020;43:579–594. - PubMed
-
- Alva-Murillo N., Ochoa-Zarzosa A., López-Meza J.E. Short chain fatty acids (propionic and hexanoic) decrease staphylococcus aureus internalization into bovine mammary epithelial cells and modulate antimicrobial peptide expression. Vet Microbiol. 2012;155:324–331. - PubMed
-
- Arora T., Sharma R., Frost G. Propionate. Anti-obesity and satiety enhancing factor? Appetite. 2011;56:511–515. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

