Systematic review and meta-analysis: Water type and temperature affect environmental DNA decay
- PMID: 35510730
- PMCID: PMC9541873
- DOI: 10.1111/1755-0998.13627
Systematic review and meta-analysis: Water type and temperature affect environmental DNA decay
Abstract
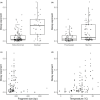
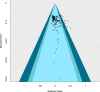
Environmental DNA (eDNA) has been used in a variety of ecological studies and management applications. The rate at which eDNA decays has been widely studied but at present it is difficult to disentangle study-specific effects from factors that universally affect eDNA degradation. To address this, a systematic review and meta-analysis was conducted on aquatic eDNA studies. Analysis revealed eDNA decayed faster at higher temperatures and in marine environments (as opposed to freshwater). DNA type (mitochondrial or nuclear) and fragment length did not affect eDNA decay rate, although a preference for <200 bp sequences in the available literature means this relationship was not assessed with longer sequences (e.g. >800 bp). At present, factors such as ultraviolet light, pH, and microbial load lacked sufficient studies to feature in the meta-analysis. Moving forward, we advocate researching these factors to further refine our understanding of eDNA decay in aquatic environments.
Keywords: aquatic eDNA; ddPCR; eDNA concentration; eDNA degradation; qPCR; species detection.
© 2022 Crown copyright. Molecular Ecology Resources published by John Wiley & Sons Ltd. This article is published with the permission of the Controller of HMSO and the Queen's Printer for Scotland.
Conflict of interest statement
We have no conflicts of interest to report.
Figures

References
-
- Akre, T. S. , Parker, L. D. , Ruther, E. , Maldonado, J. E. , Lemmon, L. , & McInerney, N. R. (2019). Concurrent visual encounter sampling validates eDNA selectivity and sensitivity for the endangered wood turtle (Glyptemys insculpta). PLoS One, 14(4), e0215586. 10.1371/journal.pone.0215586 - DOI - PMC - PubMed
-
- Andruszkiewicz, E. A. , Koseff, J. R. , Fringer, O. B. , Ouellette, N. T. , Lowe, A. B. , Edwards, C. A. , & Boehm, A. B. (2019). Modeling environmental DNA transport in the coastal ocean using Lagrangian particle tracking. Frontiers in Marine Science, 6, 477. 10.3389/fmars.2019.00477 - DOI
-
- Barnes, M. A. , & Turner, C. R. (2016). The ecology of environmental DNA and implications for conservation genetics. Conservation Genetics, 17(1), 1–17. 10.1007/s10592-015-0775-4 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

