A targeted approach to investigating immune genes of an iconic Australian marsupial
- PMID: 35510793
- PMCID: PMC9325493
- DOI: 10.1111/mec.16493
A targeted approach to investigating immune genes of an iconic Australian marsupial
Abstract
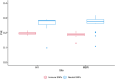
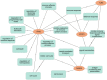
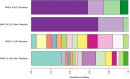
Disease is a contributing factor to the decline of wildlife populations across the globe. Koalas, iconic yet declining Australian marsupials, are predominantly impacted by two pathogens, Chlamydia and koala retrovirus. Chlamydia is an obligate intracellular bacterium and one of the most widespread sexually transmitted infections in humans worldwide. In koalas, Chlamydia infections can present as asymptomatic or can cause a range of ocular and urogenital disease signs, such as conjunctivitis, cystitis and infertility. In this study, we looked at differences in response to Chlamydia in two northern populations of koalas using a targeted gene sequencing of 1209 immune genes in addition to genome-wide reduced representation data. We identified two MHC Class I genes associated with Chlamydia disease progression as well as 25 single nucleotide polymorphisms across 17 genes that were associated with resolution of Chlamydia infection. These genes are involved in the innate immune response (TLR5) and defence (TLR5, IFNγ, SERPINE1, STAT2 and STX4). This study deepens our understanding of the role that genetics plays in disease progression in koalas and leads into future work that will use whole genome resequencing of a larger sample set to investigate in greater detail regions identified in this study. Elucidation of the role of host genetics in disease progression and resolution in koalas will directly contribute to better design of Chlamydia vaccines and management of koala populations which have recently been listed as "endangered."
Keywords: Chlamydia; GWAS; conservation genomics; koala; wildlife disease.
© 2022 The Authors. Molecular Ecology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures


References
-
- Adams‐Hosking, C. , McBride, M. F. , Baxter, G. , Burgman, M. , de Villiers, D. , Kavanagh, R. , & McAlpine, C. A. (2016). Use of expert knowledge to elicit population trends for the koala (Phascolarctos cinereus). Diversity and Distributions, 22(3), 249–262. 10.1111/ddi.12400 - DOI
-
- Ashburner, M. , Ball, C. A. , Blake, J. A. , Botstein, D. , Butler, H. , Cherry, J. M. , Davis, A. P. , Dolinski, K. , Dwight, S. S. , Eppig, J. T. , Harris, M. A. , Hill, D. P. , Issel‐Tarver, L. , Kasarskis, A. , Lewis, S. , Matese, J. C. , Richardson, J. E. , Ringwald, M. , Rubin, G. M. , & Sherlock, G. (2000). Gene ontology: tool for the unification of biology. Nature Genetics, 25(1), 25–29. 10.1038/75556 - DOI - PMC - PubMed
-
- Axelsson, E. , Ratnakumar, A. , Arendt, M.‐L. , Maqbool, K. , Webster, M. T. , Perloski, M. , Liberg, O. , Arnemo, J. M. , Hedhammar, Å. , & Lindblad‐Toh, K. (2013). The genomic signature of dog domestication reveals adaptation to a starch‐rich diet. Nature, 495(7441), 360–364. 10.1038/nature11837 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

