Broad-range metalloprotease profiling in plants uncovers immunity provided by defence-related metalloenzyme
- PMID: 35510806
- PMCID: PMC9322406
- DOI: 10.1111/nph.18200
Broad-range metalloprotease profiling in plants uncovers immunity provided by defence-related metalloenzyme
Abstract
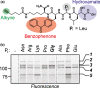
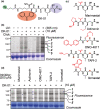
Plants encode > 100 metalloproteases representing > 19 different protein families. Tools to study this large and diverse class of proteases have not yet been introduced into plant research. We describe the use of hydroxamate-based photoaffinity probes to explore plant proteomes for metalloproteases. We detected labelling of 23 metalloproteases in leaf extracts of the model plant Arabidopsis thaliana that belong to nine different metalloprotease families and localize to different subcellular compartments. The probes identified several chloroplastic FtsH proteases, vacuolar aspartyl aminopeptidase DAP1, peroxisomal metalloprotease PMX16, extracellular matrix metalloproteases and many cytosolic metalloproteases. We also identified nonproteolytic metallohydrolases involved in the release of auxin and in the urea cycle. Studies on tobacco plants (Nicotiana benthamiana) infected with the bacterial plant pathogen Pseudomonas syringae uncovered the induced labelling of PRp27, a secreted protein with implicated metalloprotease activity. PRp27 overexpression increases resistance, and PRp27 mutants lacking metal binding site are no longer labelled, but still show increased immunity. Collectively, these studies reveal the power of broad-range metalloprotease profiling in plants using hydroxamate-based probes.
Keywords: Arabidopsis; PRp27; immunity; metalloproteases; photoaffinity labelling.
© 2022 The Authors. New Phytologist © 2022 New Phytologist Foundation.
Figures




Similar articles
-
A host-pathogen interactome uncovers phytopathogenic strategies to manipulate plant ABA responses.Plant J. 2019 Oct;100(1):187-198. doi: 10.1111/tpj.14425. Epub 2019 Jul 8. Plant J. 2019. PMID: 31148337
-
The FtsH protease heterocomplex in Arabidopsis: dispensability of type-B protease activity for proper chloroplast development.Plant Cell. 2010 Nov;22(11):3710-25. doi: 10.1105/tpc.110.079202. Epub 2010 Nov 9. Plant Cell. 2010. PMID: 21062893 Free PMC article.
-
FtsH proteases located in the plant chloroplast.Physiol Plant. 2012 May;145(1):203-14. doi: 10.1111/j.1399-3054.2011.01548.x. Epub 2012 Jan 3. Physiol Plant. 2012. PMID: 22121866 Review.
-
Absence of endo-1,4-β-glucanase KOR1 alters the jasmonate-dependent defence response to Pseudomonas syringae in Arabidopsis.J Plant Physiol. 2014 Oct 15;171(16):1524-32. doi: 10.1016/j.jplph.2014.07.006. Epub 2014 Jul 23. J Plant Physiol. 2014. PMID: 25108263
-
The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling.Mol Plant Pathol. 2017 Sep;18(7):937-948. doi: 10.1111/mpp.12457. Epub 2016 Sep 15. Mol Plant Pathol. 2017. PMID: 27399963 Free PMC article.
Cited by
-
Letter to the Editor: Cautionary Note on Ribonuclease Activity of Recombinant PR-10 Proteins.Plant Cell Physiol. 2023 Aug 17;64(8):847-849. doi: 10.1093/pcp/pcad062. Plant Cell Physiol. 2023. PMID: 37319028 Free PMC article. No abstract available.
References
-
- Baerenfaller K, Hirsch‐Hoffmann M, Svozil J, Hull R, Russenberger D, Bischof S, Lu Q, Gruissem W, Baginsky S. 2011. pep2pro: a new tool for comprehensive proteome data analysis to reveal information about organ‐specific proteomes in Arabidopsis thaliana . Integrative Biology 3: 225–237. - PubMed
-
- Cederholm SN, Bäckman HG, Pesaresi P, Leister D, Glaser E. 2009. Deletion of an organellar peptidasome PreP affects early development in Arabidopsis thaliana . Plant Molecular Biology 71: 497–508. - PubMed
-
- Christensen AB, Cho BH, Næsby M, Gregersen PL, Brandt J, Madriz‐Ordeñana K, Collinge DB, Thordal‐Christensen H. 2002. The molecular characterization of two barley proteins establishes the novel PR‐17 family of pathogenesis‐related proteins. Molecular Plant Pathology 3: 135–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

