Demographic history differences between Hispanics and Brazilians imprint haplotype features
- PMID: 35511163
- PMCID: PMC9258545
- DOI: 10.1093/g3journal/jkac111
Demographic history differences between Hispanics and Brazilians imprint haplotype features
Abstract
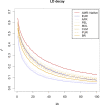
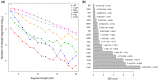
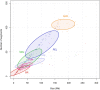
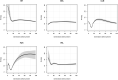
Admixture is known to greatly impact the genetic landscape of a population and, while genetic variation underlying human phenotypes has been shown to differ among populations, studies on admixed subjects are still scarce. Latin American populations are the result of complex demographic history, such as 2 or 3-way admixing events, bottlenecks and/or expansions, and adaptive events unique to the American continent. To explore the impact of these events on the genetic structure of Latino populations, we evaluated the following haplotype features: linkage disequilibrium, shared identity by descent segments, runs of homozygosity, and extended haplotype homozygosity (integrated haplotype score) in Latinos represented in the 1000 Genome Project along with array data from 171 Brazilians sampled in the South and Southeast regions of Brazil. We found that linkage disequilibrium decay relates to the amount of American and African ancestry. The extent of identity by descent sharing positively correlates with historical effective population sizes, which we found to be steady or growing, except for Puerto Ricans and Colombians. Long runs of homozygosity, a particular instance of autozygosity, was only enriched in Peruvians and Native Americans. We used simulations to account for random sampling and linkage disequilibrium to filter positive selection indexes and found 244 unique markers under selection, 26 of which are common to 2 or more populations. Some markers exhibiting positive selection signals had estimated time to the most recent common ancestor consistent with human adaptation to the American continent. In conclusion, Latino populations present highly divergent haplotype characteristics that impact genetic architecture and underlie complex phenotypes.
Keywords: IBD sharing; Latinos; ROH; diversity; haplotypes; linkage disequilibrium; population; selection.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures


References
-
- Adhikari K, Chacón-Duque JC, Mendoza-Revilla J, Fuentes-Guajardo M, Ruiz-Linares A.. The genetic diversity of the Americas. Annu Rev Genomics Hum Genet. 2017;18:277–296. - PubMed
-
- Affymetrix. BRLMM: an improved genotype calling method for the GeneChip® human mapping 500K array set [White paper]. 2006:1–18. http://tools.thermofisher.com/content/sfs/brochures/brlmm_whitepaper.pdf
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
