Analysis of evolutionary and genetic patterns in structural genes of primate lentiviruses
- PMID: 35511321
- PMCID: PMC9068864
- DOI: 10.1007/s13258-022-01257-6
Analysis of evolutionary and genetic patterns in structural genes of primate lentiviruses
Abstract
Background: Primate lentiviruses (HIV1, HIV2, and Simian immunodeficiency virus [SIV]) cause immune deficiency, encephalitis, and infectious anemia in mammals such as cattle, cat, goat, sheep, horse, and puma.
Objective: This study was designed and conducted with the main purpose of confirming the overall codon usage pattern of primate lentiviruses and exploring the evolutionary and genetic characteristics commonly or specifically expressed in HIV1, HIV2, and SIV.
Methods: The gag, pol, and env gene sequences of HIV1, HIV2, and SIV were analyzed to determine their evolutionary relationships, nucleotide compositions, codon usage patterns, neutrality, selection pressure (influence of mutational pressure and natural selection), and viral adaptation to human codon usage.
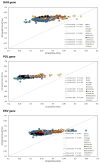
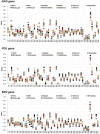
Results: A strong 'A' bias was confirmed in all three structural genes, consistent with previous findings regarding HIV. Notably, the ENC-GC3s plot and neutral evolution analysis showed that all primate lentiviruses were more affected by selection pressure than by mutation caused by the GC composition of the gene, consistent with prior reports regarding HIV1. The overall codon usage bias of pol was highest among the structural genes, while the codon usage bias of env was lowest. The virus groups showing high codon bias in all three genes were HIV1 and SIVcolobus. The codon adaptation index (CAI) and similarity D(A, B) values indicated that although there was a high degree of similarity to human codon usage in all three structural genes of HIV, this similarity was not caused by translation pressure. In addition, compared with HIV1, the codon usage of HIV2 is more similar to the human codon usage, but the overall codon usage bias is lower.
Conclusion: The origin viruses of HIV (SIVcpz_gor and SIVsmm) exhibit greater similarity to human codon usage in the gag gene, confirming their robust adaptability to human codon usage. Therefore, HIV1 and HIV2 may have evolved to avoid human codon usage by selection pressure in the gag gene after interspecies transmission from SIV hosts to humans. By overcoming safety and stability issues, information from codon usage analysis will be useful for attenuated HIV1 vaccine development. A recoded HIV1 variant can be used as a vaccine vector or in immunotherapy to induce specific innate immune responses. Further research regarding HIV1 dinucleotide usage and codon pair usage will facilitate new approaches to the treatment of AIDS.
Keywords: Codon usage pattern; Human immunodeficiency virus 1; Human immunodeficiency virus 2; Mutational pressure; Natural selection; Relative synonymous codon usage; Simian immunodeficiency virus.
© 2022. The Author(s) under exclusive licence to The Genetics Society of Korea.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
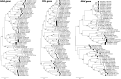
 ] SIVcol; [○] SIVcpz_gor; [●] SIVmac; [■]SIVmnd; [▼] SIVpit; [
] SIVcol; [○] SIVcpz_gor; [●] SIVmac; [■]SIVmnd; [▼] SIVpit; [ ] SIVsmm
] SIVsmm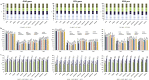



Similar articles
-
Different Patterns of Codon Usage and Amino Acid Composition across Primate Lentiviruses.Viruses. 2023 Jul 20;15(7):1580. doi: 10.3390/v15071580. Viruses. 2023. PMID: 37515266 Free PMC article.
-
Characterization of codon usage pattern and influencing factors in Japanese encephalitis virus.Virus Res. 2016 Aug 2;221:58-65. doi: 10.1016/j.virusres.2016.05.008. Epub 2016 May 14. Virus Res. 2016. PMID: 27189042
-
HIV1 V3 loop hypermutability is enhanced by the guanine usage bias in the part of env gene coding for it.In Silico Biol. 2009;9(4):255-69. In Silico Biol. 2009. PMID: 20109155
-
Human immunodeficiency virus type 2 (HIV2).Blood Rev. 1990 Sep;4(3):158-64. doi: 10.1016/0268-960x(90)90043-r. Blood Rev. 1990. PMID: 2245251 Review.
-
Origin and diversity of human retroviruses.AIDS Rev. 2014 Jan-Mar;16(1):23-34. AIDS Rev. 2014. PMID: 24584106 Free PMC article. Review.
Cited by
-
Different Patterns of Codon Usage and Amino Acid Composition across Primate Lentiviruses.Viruses. 2023 Jul 20;15(7):1580. doi: 10.3390/v15071580. Viruses. 2023. PMID: 37515266 Free PMC article.
References
-
- Apetrei C, Kaur A, Lerche NW, Metzger M, Pandrea I, Hardcastle J, Falkenstein S, Bohm R, Koehler J, Traina-Dorge V, Williams T, Staprans S, Plauche G, Veazey RS, McClure H, Lackner AA, Gormus B, Robertson DL, Marx PA. Molecular epidemiology of simian immunodeficiency virus SIVsm in US primate centers unravels the origin of SIVmac and SIVstm. J Virol. 2005;79(14):8991–9005. doi: 10.1128/Jvi.79.14.8991-9005.2005. - DOI - PMC - PubMed
-
- Deml L, Bojak A, Steck S, Graf M, Wild J, Schirmbeck R, Wolf H, Wagner R. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J Virol. 2001;75(22):10991–11001. doi: 10.1128/JVI.75.22.10991-11001.2001. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
