Idiosyncratic epistasis leads to global fitness-correlated trends
- PMID: 35511982
- PMCID: PMC10124986
- DOI: 10.1126/science.abm4774
Idiosyncratic epistasis leads to global fitness-correlated trends
Abstract
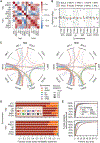
Epistasis can markedly affect evolutionary trajectories. In recent decades, protein-level fitness landscapes have revealed extensive idiosyncratic epistasis among specific mutations. By contrast, other work has found ubiquitous and apparently nonspecific patterns of global diminishing-returns and increasing-costs epistasis among mutations across the genome. Here, we used a hierarchical CRISPR gene drive system to construct all combinations of 10 missense mutations from across the genome in budding yeast and measured their fitness in six environments. We show that the resulting fitness landscapes exhibit global fitness-correlated trends but that these trends emerge from specific idiosyncratic interactions. We thus provide experimental validation of recent theoretical work arguing that fitness-correlated trends can emerge as the generic consequence of idiosyncratic epistasis.
Figures


References
-
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, Pelechano V, Styles EB, Billmann M, van Leeuwen J, van Dyk N, Lin Z-Y, Kuzmin E, Nelson J, Piotrowski JS, Srikumar T, Bahr S, Chen Y, Deshpande R, Kurat CF, Li SC, Li Z, Usaj MM, Okada H, Pascoe N, San Luis B-J, Sharifpoor S, Shuteriqi E, Simpkins SW, Snider J, Suresh HG, Tan Y, Zhu H, Malod-Dognin N, Janjic V, Przulj N, Troyanskaya OG, Stagljar I, Xia T, Ohya Y, Gingras A-C, Raught B, Boutros M, Steinmetz LM, Moore CL, Rosebrock AP, Caudy AA, Myers CL, Andrews B, Boone C, A global genetic interaction network maps a wiring diagram of cellular function. Science. 353 (2016), doi: 10.1126/science.aaf1420. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

