A comparative transcriptomics and eQTL approach identifies SlWD40 as a tomato fruit ripening regulator
- PMID: 35512210
- PMCID: PMC9434188
- DOI: 10.1093/plphys/kiac200
A comparative transcriptomics and eQTL approach identifies SlWD40 as a tomato fruit ripening regulator
Erratum in
-
Correction to: A comparative transcriptomics and eQTL approach identifies SlWD40 as a tomato fruit ripening regulator.Plant Physiol. 2023 May 31;192(2):1657. doi: 10.1093/plphys/kiad190. Plant Physiol. 2023. PMID: 36995995 Free PMC article. No abstract available.
Abstract
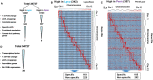
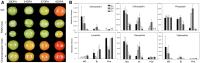
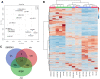
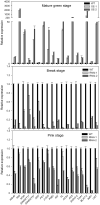
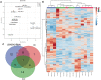
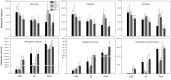
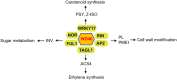
Although multiple vital genes with strong effects on the tomato (Solanum lycopersicum) ripening process have been identified via the positional cloning of ripening mutants and cloning of ripening-related transcription factors (TFs), recent studies suggest that it is unlikely that we have fully characterized the gene regulatory networks underpinning this process. Here, combining comparative transcriptomics and expression QTLs, we identified 16 candidate genes involved in tomato fruit ripening and validated them through virus-induced gene silencing analysis. To further confirm the accuracy of the approach, one potential ripening regulator, SlWD40 (WD-40 repeats), was chosen for in-depth analysis. Co-expression network analysis indicated that master regulators such as RIN (ripening inhibitor) and NOR (nonripening) as well as vital TFs including FUL1 (FRUITFUL1), SlNAC4 (NAM, ATAF1,2, and CUC2 4), and AP2a (Activating enhancer binding Protein 2 alpha) strongly co-expressed with SlWD40. Furthermore, SlWD40 overexpression and RNAi lines exhibited substantially accelerated and delayed ripening phenotypes compared with the wild type, respectively. Moreover, transcriptome analysis of these transgenics revealed that expression patterns of ethylene biosynthesis genes, phytoene synthase, pectate lyase, and branched chain amino transferase 2, in SlWD40-RNAi lines were similar to those of rin and nor fruits, which further demonstrated that SlWD40 may act as an important ripening regulator in conjunction with RIN and NOR. These results are discussed in the context of current models of ripening and in terms of the use of comparative genomics and transcriptomics as an effective route for isolating causal genes underlying differences in genotypes.
© The Author(s) 2022. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures



Comment in
-
The time is ripe for eQTLs: Transcriptomic identification of a tomato fruit ripening regulator.Plant Physiol. 2022 Aug 29;190(1):182-184. doi: 10.1093/plphys/kiac287. Plant Physiol. 2022. PMID: 35703978 Free PMC article. No abstract available.
Similar articles
-
A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation.Plant Cell Physiol. 2014 Jan;55(1):119-35. doi: 10.1093/pcp/pct162. Epub 2013 Nov 20. Plant Cell Physiol. 2014. PMID: 24265273
-
A tomato NAC transcription factor, SlNAM1, positively regulates ethylene biosynthesis and the onset of tomato fruit ripening.Plant J. 2021 Dec;108(5):1317-1331. doi: 10.1111/tpj.15512. Epub 2021 Nov 2. Plant J. 2021. PMID: 34580960
-
The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening.Plant Cell. 2012 Nov;24(11):4437-51. doi: 10.1105/tpc.112.103283. Epub 2012 Nov 6. Plant Cell. 2012. PMID: 23136376 Free PMC article.
-
The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: a review.Plant Mol Biol. 2021 Jun;106(3):223-238. doi: 10.1007/s11103-021-01128-w. Epub 2021 Feb 25. Plant Mol Biol. 2021. PMID: 33634368 Review.
-
Use of genomics tools to isolate key ripening genes and analyse fruit maturation in tomato.J Exp Bot. 2002 Oct;53(377):2023-30. doi: 10.1093/jxb/erf057. J Exp Bot. 2002. PMID: 12324526 Review.
Cited by
-
Genome-Wide Identification of the WD40 Gene Family in Tomato (Solanum lycopersicum L.).Genes (Basel). 2023 Jun 15;14(6):1273. doi: 10.3390/genes14061273. Genes (Basel). 2023. PMID: 37372453 Free PMC article. Review.
-
Identification of candidate genes that regulate the trade-off between seedling cold tolerance and fruit quality in melon (Cucumis melo L.).Hortic Res. 2023 May 9;10(7):uhad093. doi: 10.1093/hr/uhad093. eCollection 2023 Jun. Hortic Res. 2023. PMID: 37416729 Free PMC article.
-
The time is ripe for eQTLs: Transcriptomic identification of a tomato fruit ripening regulator.Plant Physiol. 2022 Aug 29;190(1):182-184. doi: 10.1093/plphys/kiac287. Plant Physiol. 2022. PMID: 35703978 Free PMC article. No abstract available.
-
Genomic insights into the domestication and genetic basis of yield in papaya.Hortic Res. 2025 Feb 17;12(5):uhaf045. doi: 10.1093/hr/uhaf045. eCollection 2025 May. Hortic Res. 2025. PMID: 40236729 Free PMC article.
-
Prospects of microgreens as budding living functional food: Breeding and biofortification through OMICS and other approaches for nutritional security.Front Genet. 2023 Jan 25;14:1053810. doi: 10.3389/fgene.2023.1053810. eCollection 2023. Front Genet. 2023. PMID: 36760994 Free PMC article. Review.
References
-
- Alseekh S, Ofner I, Pleban T, Tripodi P, Di Dato F, Cammareri M, Mohammad A, Grandillo S, Fernie AR, Zamir D (2013) Resolution by recombination: breaking up Solanum pennellii introgressions. Trends Plant Sci 18: 536–538 - PubMed
-
- Ballester AR, Molthoff J, de Vos R, Hekkert B, Orzaez D, Fernandez-Moreno JP, Tripodi P, Grandillo S, Martin C, Heldens J, et al. (2010) Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol 152: 71–84 - PMC - PubMed
-
- Bartley GE, Scolnik PA (1993) cDNA cloning, expression during development, and genome mapping of PSY2, a second tomato gene encoding phytoene synthase. J Biol Chem 268: 25718–25721 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

