A coiled-coil protein associates Polycomb Repressive Complex 2 with KNOX/BELL transcription factors to maintain silencing of cell differentiation-promoting genes in the shoot apex
- PMID: 35512211
- PMCID: PMC9338815
- DOI: 10.1093/plcell/koac133
A coiled-coil protein associates Polycomb Repressive Complex 2 with KNOX/BELL transcription factors to maintain silencing of cell differentiation-promoting genes in the shoot apex
Abstract
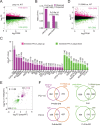
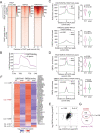
Polycomb repressive complex 2 (PRC2), which mediates the deposition of H3K27me3 histone marks, is important for developmental decisions in animals and plants. In the shoot apical meristem (SAM), Three Amino acid Loop Extension family KNOTTED-LIKE HOMEOBOX /BEL-like (KNOX/BELL) transcription factors are key regulators of meristem cell pluripotency and differentiation. Here, we identified a PRC2-associated coiled-coil protein (PACP) that interacts with KNOX/BELL transcription factors in rice (Oryza sativa) shoot apex cells. A loss-of-function mutation of PACP resulted in differential gene expression similar to that observed in PRC2 gene knockdown plants, reduced H3K27me3 levels, and reduced genome-wide binding of the PRC2 core component EMF2b. The genomic binding of PACP displayed a similar distribution pattern to EMF2b, and genomic regions with high PACP- and EMF2b-binding signals were marked by high levels of H3K27me3. We show that PACP is required for the repression of cell differentiation-promoting genes targeted by a rice KNOX1 protein in the SAM. PACP is involved in the recruitment or stabilization of PRC2 to genes targeted by KNOX/BELL transcription factors to maintain H3K27me3 and gene repression in dividing cells of the shoot apex. Our results provide insight into PRC2-mediated maintenance of H3K27me3 and the mechanism by which KNOX/BELL proteins regulate SAM development.
� American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures






Comment in
-
PACP recruits PRC2 to TALE targets.Plant Cell. 2022 Jul 30;34(8):2821-2822. doi: 10.1093/plcell/koac159. Plant Cell. 2022. PMID: 35666578 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

